ForenSeq Kintelligence HT Kit
Cat no. / ID. V16000190
Features
- Targets 10,230 SNP markers explicitly curated for identification of unidentified persons and remains
- Overcomes challenges with bone samples with optimized PCR buffer
- Unique dual index (UDI) plate facilitates simple manual or automated transfer of the index adapters
- Up to 12 postmortem and 36 antemortem samples sequenced simultaneously to reach third degree relatives
Product Details
ForenSeq Kintelligence HT is the first targeted sequencing workflow designed for unidentified persons and remains or large-scale kinship projects. Targeting 10,230 SNP markers specifically curated for kinship determination, the assay is optimized to work with challenging samples such as bones. Leveraging the unique dual index (UDI) plate allows the library preparation workflow processing of up to 96 postmortem (PM) or antemortem (AM) samples before moving on to sequencing.
The ForenSeq Kintelligence HT workflow enables a higher level of multiplexing that balances throughput with the closer kinship determinations typically required for unidentified persons and remains. Detect at least third-degree kinship with 100% sensitivity and specificity when sequencing up to 12 postmortem samples or 36 antemortem samples per MiSeq FGx run. Analyze data in the Universal Analysis Software (UAS) using the preconfigured ForenSeq Kintelligence HT Analysis Method. Utilize the new UAS database exclusively for Kintelligence HT featuring local database storage, kinship comparison of postmortem to antemortem samples, and likelihood ratios for relationship comparisons.
Performance
The ForenSeq Kintelligence HT kit is the first forensic assay to combine high throughput targeted SNP sequencing and local kinship analysis database for the large-scale identification of human remains. The fully integrated workflow includes optimized chemistry for highly degraded and contaminated samples, higher level multiplexing that balances throughput with closer kinship determinations, and a fit-for-purpose analysis software and kinship database that can be used on a local database to maintain the privacy of the families concerned.
ForenSeq Kintelliegnce HT has optimized workflows for both postmortem (PM) and antemortem samples (AM) with sequencing capabilities up to 12 PM libraries and 36 AM libraries obtaining at least third degree relationship.
High throughput, fit for purpose workflow:
- Kinship power beyond STRs
Reliably determine relationships out to the third degree, even with degraded samples. - Sensitive solution for low input, low quality samples
Sequence compromised samples on a forensically validated system for easily accessible results. - A local database for kinship analysis
Pair two familiar workflows, ForenSeq and GEDmatch PRO algorithms for kinship testing
| Feature | Details | |
|---|---|---|
| Kit configuration | 96 reactions | |
| Number of SNPs | 10,230 | |
| Mean amplicon size | < 150 bp | |
| Total library prep time | 10 hours | |
| Hands-on library prep time | < 3 hours | |
| Sequencing time | 28 hours | |
| Sample type tested | Blood, bone, buccal swabs, rooted hair, teeth, and semen | |
| Antemortem samples | Postmortem samples | |
| Recommended input | 1 ng | 500pg |
| Multiplexing capacity | 36 libraries per run | 12 libraries per run |
Principle
A robust PCR and primer design enables a single SNP multiplex, maximising kinship power. The small average amplicon size of < 150 bp improves amplification efficiency and facilitates recovery and analysis of degraded DNA. Additionally, the ForenSeq Kintelligence HT Kit includes 96 unique dual index (UDI) adapters, which attach unique identifying sequences to each end of the library for optimum data recovery.
ForenSeq Kintelligence HT queries 10,230 SNPs carefully selected to maximise kinship power.
| Category | Number of SNPs | Percentage of total |
| Ancestry SNPs | 56 | 0,5% |
| Identity SNPs | 94 | 1% |
| Kinship SNPs | 9867 | 96% |
| Phenotype SNPs* | 22 | 0,2% |
| X-SNPs | 106 | 1,2% |
| Y-SNPs | 85 | 0,9% |
*Two SNPs overlap the ancestry and phenotype categories and are counted in the phenotype category only.
The ForenSeq Kintelligence HT Kit utilizes the same SNPs found in the original ForenSeq Kintelligence Kit, including the 9867 kinship SNPS, as well as all biogeographical ancestry, identity and phenotype SNPs validated for investigative lead generation. Also included are informative Y-SNPs and X-SNPs to help with lineage and biological sex determination, respectively.
These SNPs overlap with relevant markers in the Illumina Infinium CytoSNP-850K BeadChip and Infinium Global Screening Array and are cross referenced against the Genome Aggregation Database (gnomAD) v3.0 and Single Nucleotide Polymorphism database (dbSNP) v151 for robust performance across global populations. Importantly, ForenSeq Kintelligence HT excludes the SNPs with known medical associations or low minor allele frequencies to limit privacy concerns and protect genetic health data.
An optimized buffer system tolerates a range of inhibitors commonly found in forensic samples, such as calcium, indigo and humic acid, generating a > 98% call rate. Assay design provides a high level of process integrity and data quality so you can trust the analysis of even the most challenging forensic samples.
Procedure
ForenSeq Kintelligence HT inputs are compatible with DNA extraction methods common in forensic laboratories. Library preparation leverages the same chemistry backbone that is foundational to the ForenSeq library prep portfolio. ForenSeq Kintelligence HT reagents enable preparation of up to 96 dual-indexed, human-specific libraries in 10 hours, with < 3 hours of hands-on time. Pooling libraries recommendation for sequencing are 12 PM samples or 36 AM samples.
Sequencing is completed in 28 hours and analysis in 1 hour, including upload to the local UAS database for kinship and likelihood analysis. The fully integrated NGS workflow delivers results. In addition to providing streamlined library prep that includes normalization, the ForenSeq chemistry offers a high degree of flexibility for sample sources such as buccal swabs, blood, bones and teeth. The 96-reaction kit includes master mixes for amplification, purification beads for cleanup and is supplied with straightforward protocols, featuring six safe stopping points.
Applications
The ForenSeq Kintelligence HT Kit is the first forensic assay to combine high throughput targeted SNP sequencing and local kinship analysis for the identification of human remains in mass disaster or large-scale kinship projects. The fully integrated workflow includes optimized chemistry for highly degraded and contaminated samples and fit for purpose analysis software with a local kinship database. Library multiplexing recommendations balance throughput with the degree of relationship requirements.
Software
As with all human identification scenarios, protection of genetic data from victims and families is important. Database access should be controlled for legislative and ethical reasons. The ForenSeq Kintelligence HT workflow includes the ability to create a private database within the UAS and built-in analysis tools repurposed from GEDmatch PRO to maintain sensitive information within a forensic environment.
Samples processed with the ForenSeq Kintelligence HT Kit are analzyed using the UAS Kintelligence HT analysis module where users can view data and make edits if needed. Sample results are then accessable in the UAS database where kinship comparisons between two samples can be made utilizing the Query feature. When a user initiates a Query, an unknown sample(s) and a reference sampe(s) are compared in a pairwise manner. A range of metrics are displayed including shared cM, longest segment and kingship coefficient, allowing the user to make a determination of pedigree. Likelihood ratios are calculated for the compared samples when a specific relationship has been selected.
Services
We offer superior support across the entire workflow, from library prep to sequencing to analysis. Our experienced team provides comprehensive service coverage for your equipment and software, validation plans and implementation guidance so you can operationalize your workflows with ease.
Supporting data and figures
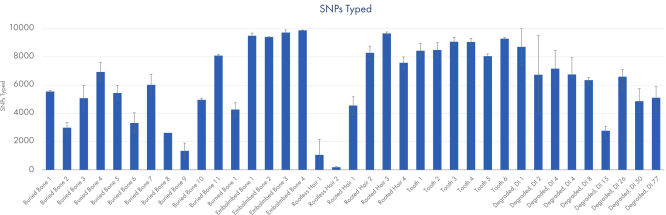
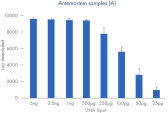
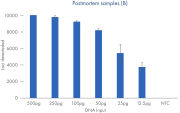
Call rates for different degraded sample types typically encountered in cases of unidentified human remains. Data shown from runs sequenced at a plexity of 12 libraries.
The ForenSeq Kintelligence HT Kit was tested on a range of sample types typically encountered when testing human remains: interred bones, embalmed bones, teeth and hair. An artificially degraded DNA series (extracted from blood) purchased from Innogenomics technologies (New Orleans LA, USA) was also tested. The ForenSeq Kintelligence HT Kit delivered high performance across all sample types. Call rates of over 7,000 SNPs were obtained for the majority of samples except for the rootless hair which is consistent with expected recovery rates at the recommended sample plexity.