QIAseq xHYB CGP DNA Panel (24)
Cat no. / ID. 333122
Features
- Covers 720+ genes, detecting SNVs, Indels, CNVs, MSI, TMB and RNA fusions
- Analyze DNA & RNA variants from one sample by pairing with QIAseq Multimodal DNA/RNA Lib Kit
- Captures challenging regions, including regions with high GC content
- Compatible with multiple sample types, including FFPE and liquid biopsy samples
- Customize panels or expand coverage with additional probes
Product Details
QIAseq xHYB CGP Panels are expertly curated for comprehensive genomic profiling (CGP). These panels target key genomic regions, incorporating genes with actionable and interpretable variants sourced from the Human Somatic Mutation Database (HSMD) using tier 1 and tier 2 actionability filters. They also align with clinical practice guidelines recommended by multiple organizations, ensuring high coverage of critical regions across more than 720 genes. The optimized probe design ensures efficient capture of challenging regions, including high GC-content areas, providing reliable data from the most challenging samples, such as FFPE and cfDNA samples.
QIAseq xHYB CGP Panels can be combined with QIAseq Multimodal DNA/RNA Library Kit for comprehensive genomic profiling from a single total nucleic acid sample.
Choose the right CGP panel for your research needs:
| Panel name | Specification |
|---|---|
| QIAseq xHYB CGP DNA Panel | Detects SNVs and indels in 724 oncogenes and tumor suppressor genes; supports TMB and MSI analyses |
| QIAseq xHYB CGP RNA Panel | Detects fusions, splicing and exon-skipping in 274 genes |
| QIAseq xHYB DNA Fusion and MSI+ Panel | In the absence of RNA, detects DNA fusions in 50 genes; targets >4000 MSI markers for optimized MSI analysis; designed for cfDNA samples and can be used as a stand-alone panel or as a spike-in for QIAseq xHYB CGP DNA Panel |
Customize the CGP panel to your variants of interest:
- Target specific regions of interest with fully custom panels
- Seamlessly expand existing QIAseq xHYB CGP Panel coverage
- Boost coverage by adding extra probes for deeper biomarker insights
Performance
High coverage uniformity and sensitivity
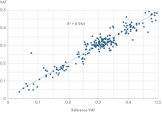
QIAseq xHYB CGP Panels deliver exceptional coverage uniformity, ensuring >99% of target bases are covered at ≥20x, regardless of GC content. This enables highly sensitive variant detection, with >95% combined sensitivity and >99.995% specificity for small variants, while minimizing dropout.
Fast turnaround
With hybridization times as short as 30 minutes and full automation compatibility, QIAseq xHYB CGP Panels provide fast, high-quality reproducible results. When paired with the QIAseq Multimodal DNA/RNA Library Kit, they enable comprehensive genomic profiling from a single total nucleic acid sample. The streamlined workflow integrates unique molecular indices (UMIs) for accuracy, delivering sequencing-ready libraries in just 1.5 days.
Sample type flexibility
QIAseq xHYB CGP Panels enable comprehensive genomic profiling from both FFPE and liquid biopsy samples, ensuring accurate and actionable insights for translational research applications.
- For FFPE samples, combining the DNA and RNA panels provides a complete view of genomic alterations, including SNVs, indels, CNVs, TMB, MSI and RNA fusions
- For liquid biopsy samples, pairing the DNA panel with the Fusion and MSI+ panel enhances sensitivity for detecting oncogenic fusions in 50 genes and enhances MSI detection in low-input cfDNA
| Features | Specifications |
|---|---|
| Variants called | SNVs, InDels, CNVs, fusions and splice variants |
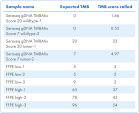
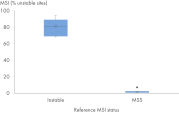
| Genomic signatures called | TMB, MSI |
| Total turnaround time | 1.5 days from nucleic acid to sequencing-ready library |
| Sample multiplexing for hybrid capture | Up to 8-plex |
| Species | Human |
| Specialized sample types | FFPE, cfDNA |
| Nucleic acid input |
40 ng DNA, 40–80 ng RNA Note: Input reduction may affect the detection of variants below the 5% VAF limit of detection |
| Analytical sensitivity | >95% (small variants, 5% VAF) |
| Analytical specificity | >99.995% (small variants, 5% VAF) |
|
Recommended raw reads (in million) Based on 2x150 bp |
75 million for FFPE DNA, 25 million for FFPE RNA |
| Recommended data analysis software |
QIAGEN CLC Genomics Workbench for secondary analysis QIAGEN Clinical Insight for tertiary analysis |
| Total assay time | 5 days from sample to tertiary report |
Principle
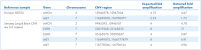
QIAseq xHYB CGP Panels use a hybridization capture-based target enrichment approach to specifically enrich curated sequences of the human genome associated with known variants from indexed whole genome or whole transcriptome libraries. The flexible workflow allows simultaneous hybrid capture from up to 8 samples with as little as 200 ng input per library.
Once the probes hybridize with their targets, the probe-target hybrids bind to streptavidin-coated magnetic beads through interaction with the biotin label on the probes. A magnet is then used to keep these beads with the probe-target hybrids against the side of the tube while the unbound DNA is washed away, reducing off-target effects. The targets are then amplified and prepared for sequencing.
Procedure
QIAseq Multimodal DNA/RNA Library Kit is recommended for whole genome and whole transcriptome (WGS/WTS) library prep prior to hybrid capture with the QIAseq xHYB CGP Panels. Other WGS/WTS library prep kits are also compatible, as long as the libraries are prepared without modifications with biotin. QIAseq Multimodal DNA/RNA Library Kit starts with a single total nucleic acid sample and produces DNA and RNA libraries for further target enrichment, with safe stopping points to proceed with DNA only, RNA only or both DNA and RNA. This is followed by target enrichment by hybrid capture using the QIAseq xHYB CGP workflow followed by sequencing.
The resulting FASTQ files are uploaded into the QIAseq xHYB CGP workflow within the QIAGEN CLC Genomics Workbench for filtering, read mapping and variant calling. The VCF output is uploaded into QIAGEN Clinical Insight – Interpret (QCI-I), enabling a variant filtering cascade that facilitates the prioritization of variants for evidence-based interpretation.
Applications
QIAseq xHYB CGP Panels provide comprehensive genomic insights to advance cancer research:
- Assess TMB and MSI to optimize immunotherapy outcomes
- Monitor shifts in mutations and fusions over time to study tumor heterogeneity and resistance
- Consolidate comprehensive genomic data into one assay for streamlined research efficiency
- Detect actionable alterations to validate targeted treatment efficacy
Supporting data and figures
Workflow principle of QIAseq xHYB CGP Panels
Isolated DNA and RNA are mixed in a single tube, or isolated total nucleic acids are used. In the first step, DNA is enzymatically fragmented, end-repaired and A-tailed. rRNA removal is performed by adding QIAseq FastSelect® –rRNA reagent. In the second step, RNA is specifically polyadenylated. The DNA adapter (optional use of UMIs) is ligated to DNA in step 3. The sample is split into two tubes. The DNA library is indexed with the UDIs provided. The RNA is then reverse transcribed, and the RNA library indexed using the UDIs provided. Both libraries are hybrid captured for target enrichment using QIAseq xHYB CGP Panels.