Taq DNA Polymerase (250 U)
Cat no. / ID. 201203
Features
- Choice of formats for convenience and ease of handling
- QIAGEN PCR Buffer for minimal optimization
- Q-Solution for amplification of GC-rich templates
- Additional ready-to-load PCR buffer for faster handling
Product Details
Taq DNA Polymerase is a replicative polymerase derived from the thermophilic eubacterium Thermus aquaticus. Thermostable activity at temperatures above 70°C makes the enzyme suitable for standard and specialized PCR amplification applications. QIAGEN Taq DNA Polymerase is supplied with PCR Buffer specially formulated for fast setup with minimal optimization of PCR parameters, Q-Solution to facilitate amplification of "difficult" (e.g., GC-rich) templates and the additional time-saving advantage of CoralLoad PCR Buffer with two gel-tracking dyes to enable immediate loading of PCR products.
QIAGEN Taq DNA Polymerase is also available in Taq PCR Core Kit that includes dNTPs. If you prefer a Master Mix format, including all needed components, go to Taq PCR Master Mix Kit.
The source of Taq DNA polymerase, the thermophilic eubacterium Thermus aquaticus, was first identified in 1966, living and thriving at about 75°C in the waters of a hot spring in Yellowstone National Park. Taq DNA polymerase has a temperature optimum of 72°C and does not denature at 95°C. This intrinsic thermostability makes Taq DNA polymerase suitable for standard and specialized PCR amplification applications based on its core competence to maintain relatively high catalytic activity and stability after multiple rounds of thermal cycling at high temperatures.
Taq DNA polymerase has 5’→3’ exonuclease activity to remove RNA primers but lacks 3’→5’ exonuclease “proofreading” activity. Many DNA polymerases perform highly accurate DNA synthesis even in the absence of exonucleolytic proofreading. In nature, Thermus aquaticus compensates for the lack of proofreading with a mismatch repair (MMR) system including a MutS homolog that plays a crucial role in correcting replication errors. Taq DNA polymerase performs best when amplifying DNA fragments <2 kb but can amplify longer fragments efficiently under defined reaction conditions — dNTP concentration, pH and the concentration of MgCl2 relative to the total concentration of dNTPs present. Under these conditions, an error rate for Taq DNA polymerase per nucleotide polymerized at 70°C can be achieved as low as 10-5 for base substitution errors and 10-6 for frameshift errors.
The non-template-dependent terminal transferase activity inherent in Taq DNA polymerase and other nonproofreading DNA polymerases provides a highly efficient method to clone PCR products. Taq DNA polymerase adds a single, unpaired residue, preferentially an adenosyl residue, to each 3'-end of a double-stranded amplified product (extra A addition). This property is an advantage for the TA-cloning strategy but may present drawbacks when Taq DNA polymerase is used for microsatellite genotyping analysis.
QIAGEN Taq DNA Polymerase is designed to deliver fast, reliable PCR-related workflows that are easily optimized for efficient amplification performance and reduced error rates.
If you are looking for a DNA polymerase with higher fidelity or longer range than Taq DNA Polymerase, explore our long range or higher fidelity enzymes and mixes. Long range PCR provides highly sensitive and specific long-range amplification for up to 30 kb using any DNA or cDNA template. High fidelity enzymes and master mixes confer higher fidelity, speed and performance compared to standard Taq DNA Polymerase.
Performance
Taq DNA Polymerase outperforms Taq enzyme systems from other suppliers and delivers efficient PCR performance, including for low-copy, longer or difficult targets, over a wide range of conditions without the need for time-consuming optimization.
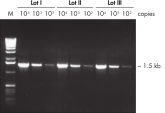
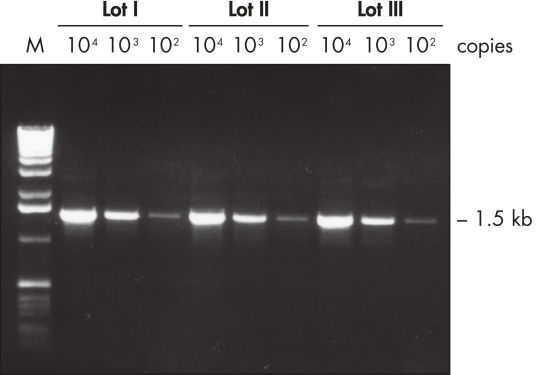
Lot-to-lot reproducibility with low-copy targets.
Specificity and reproducibility of PCR with QIAGEN Taq DNA Polymerase were demonstrated when a low-copy target was amplified reproducibly from human genomic DNA by three different lots of the enzyme (see figure " Lot-to-lot reproducibility"). A fragment of the single-copy gene for cystic fibrosis was amplified with three different lots of QIAGEN Taq DNA Polymerase under standard conditions from 30 ng, 3 ng, and 300 pg human genomic DNA corresponding to 104, 103, and 102 copies of target template, respectively. No differences were seen among the lots when equal volumes of the PCR product were analyzed on a 1% agarose gel.
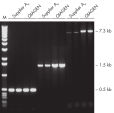
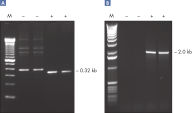
Specific amplification of PCR products longer than 5 kb.
QIAGEN Taq DNA Polymerase with PCR Buffer demonstrated greater capacity than a rival enzyme to successfully amplify a target longer than 5 kb (see figure " Specific amplification of long PCR products"). Three differently sized products from human genomic DNA were amplified in-house in parallel using either QIAGEN Taq DNA Polymerase and PCR Buffer under standard conditions, or Taq DNA polymerase and buffer from another supplier following the manufacturer’s instructions. Results of amplifications of successively longer amplicons analyzed on a 1% agarose gel show an increasingly wide disparity of efficiency between QIAGEN Taq DNA Polymerase and Taq from the other supplier. Product concentrations from both systems were similar for a short amplicon (0.5 kb) while QIAGEN Taq DNA Polymerase outperformed the rival Taq about two-fold for a slightly longer amplicon (1.5 kb). There was a marked difference (about 10-fold) between the two enzyme systems when the target was 7.3 kb in length.
Wide annealing temperature window (50–60°C) with no optimization.
PCR amplification with QIAGEN Taq DNA Polymerase and PCR Buffer was more tolerant of a wide range of annealing temperatures than enzyme from another supplier. Optimization of annealing temperature was not required to ensure efficient amplification (see figure " A. Wide annealing-temperature window"). A fragment of the single-copy gene for cystic fibrosis was amplified from human genomic DNA in-house using either QIAGEN Taq DNA Polymerase and PCR Buffer under standard conditions, or Taq DNA polymerase and buffer from another supplier following the manufacturer’s instructions. PCR amplifications were conducted in parallel without additional optimization at annealing temperatures with 2-degree increments between 50°C and 60°C. Equal volumes of the PCR products were analyzed on a 1% agarose gel.
Tolerance of variable magnesium concentration (1.5–4.0 mM) with no optimization.
PCR amplification with QIAGEN Taq DNA Polymerase and PCR Buffer was more tolerant of a wide range of magnesium ion concentrations than an enzyme from another supplier. Optimization of Mg2+ was not required to ensure efficient amplification (see figure " B. Tolerance of variable magnesium concentration”). A fragment of the single-copy prion gene was amplified from human genomic DNA in-house using either QIAGEN Taq DNA Polymerase and PCR Buffer under standard conditions, or Taq DNA polymerase and buffer from another supplier following the manufacturer’s instructions. PCR amplifications were conducted in parallel without additional optimization using Mg2+ concentrations with increments of 0.5 mM between 1.5 mM and 4.0 mM. Equal volumes of the PCR products were analyzed on a 1% agarose gel.
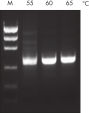
Tolerance of different primer Tm values (>25°C) with no optimization.
PCR amplification with Taq DNA Polymerase and PCR Buffer was tolerant of very different primer pair Tm values (>25°C difference). Matching of Tm values and optimization of annealing temperature were not required to ensure efficient amplification (see figure: “ Tolerance of different primer Tm values”). The human single-copy cystic fibrosis gene was amplified under standard conditions with Taq DNA Polymerase and PCR Buffer and the same primers using three different annealing temperatures — 55°C, 60°C and 65°C. Primers employed were a 22mer with a Tm of 57.5°C (GC content: 54.5%) and a 32mer with a Tm of 85.2°C (GC content: 78%). Analysis of 10% of each 100 µL reaction on a 1% agarose gel showed consistent and reliable amplification of the target under these suboptimal PCR conditions.
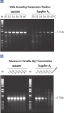
Amplification of difficult templates assisted by Q-Solution.
Q-Solution is included with Taq DNA Polymerase and Taq PCR Core Kit. This reagent modifies the melting behavior of DNA and facilitates amplification of GC-rich templates or templates with a high degree of secondary structure (see figure: " Amplification of difficult templates with Q-Solution”). Amplification in duplicate was conducted with Taq DNA Polymerase and PCR Buffer in the absence (-) or presence (+) of 1x Q-Solution using two different primer-template systems for (A) the human angiotensin receptor II gene and (B) the mouse protein kinase C gene. Analysis of equal volumes of the PCR products on a 1% agarose gel clearly showed the advantage of adding Q-Solution to these PCR reactions.
Shipping and storage.
Taq DNA Polymerase and Taq PCR Core Kit are shipped on dry ice but retain full activity at room temperature (15–25°C) for 2 weeks.
Taq DNA Polymerase and the Taq PCR Core Kit, including buffers and reagents, should be stored immediately upon receipt at –20°C in a constant-temperature freezer. When stored under these conditions and handled correctly, these products can be kept at least until the expiration date without showing any reduction in performance.
Taq DNA Polymerase enzyme specifications
| Features | Specifications |
| Concentration | 5 units/µL |
| Recombinant enzyme | Yes |
| Substrate analogs | dNTP, ddNTP, dUTP, biotin-11-dUTP, DIG-11-dUTP, fluorescent-dNTP/ddNTP |
| Extension rate | 2–4 kb/min at 72°C |
| Half life | 10 min at 97°C; 60 min at 94°C |
| Amplification efficiency | ≥105 fold |
| 5’→3’ exonuclease activity | Yes |
| Extra A addition | Yes |
| 3’→5’ exonuclease proofreading activity | No |
| Contaminating nucleases | No |
| Contaminating RNases | No |
| Contaminating proteases | No |
| Self-priming activity | No |
Principle
Taq DNA Polymerase is a high-quality recombinant enzyme produced by QIAGEN. This enzyme is suitable for routine PCR (e.g., probes, gene expression analysis and cloning) as well as specialized PCR applications (e.g., PCR-based DNA fingerprinting and differential display to identify genes differentially expressed between two or more cell or tissue samples).
Taq DNA Polymerase is used for PCR in combination with PCR Buffer or CoralLoad PCR Buffer for reproducible results without the need for time-consuming optimization. These are specialized PCR buffers developed to save time and effort by reducing the need for optimization of individual primer–template systems. Q-Solution is available to facilitate amplification of GC-rich templates or templates with a high degree of secondary structure.
PCR Buffer
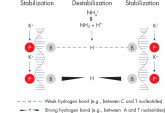
QIAGEN PCR Buffer minimizes the need for PCR optimization and saves time and effort by eliminating extra steps taken to establish an ideal annealing temperature or Mg2+ concentration (see figure: " A. Wide annealing temperature window. B. Tolerance of variable magnesium concentration"). A balanced combination of KCl and (NH4)2SO4 in PCR Buffer results in primer-annealing conditions that are more stringent over a wider range of temperatures and Mg2+ concentrations than those provided by conventional PCR buffers. The combined effect of the NH4+ and K+ cations maintains the high ratio of specific to nonspecific primer–template binding during the annealing step of every PCR cycle (see figure: "NH4+ and K+ cations in QIAGEN PCR Buffer increase specific primer annealing").
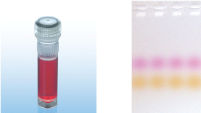
CoralLoad PCR Buffer
CoralLoad PCR Buffer provides the same high PCR specificity and minimal reaction optimization as conventional PCR Buffer with two additional advantages. For enhanced convenience, the buffer contains orange and red marker dyes that improve pipetting visibility and enable direct gel loading of PCR products for estimation of DNA migration distance and optimization of agarose gel run time (see figure " CoralLoad PCR Buffer").
Q-Solution
Q-Solution modifies the melting behavior of DNA. Use of this reagent may improve suboptimal PCR and enable amplification of GC-rich templates or templates with a high degree of secondary structure (see figure: " Amplification of difficult templates with Q-Solution"). Unlike DMSO and other PCR additives, Q-Solution is used at a defined working concentration with any primer–template system and is not toxic.
Procedure
PCR amplification
The protocol for PCR amplification with Taq DNA Polymerase used in combination with PCR Buffer or CoralLoad PCR Buffer follows straightforward guidelines (see Taq PCR Handbook). Whereas some optimal reaction conditions, such as incubation times and amount of template DNA, may vary and should be individually determined, reproducible results are possible with this system without the need for optimization of other parameters such as annealing temperature, magnesium concentration and Tm values of primers.
If CoralLoad PCR Buffer is used, the PCR product can be directly loaded onto an agarose gel without prior addition of a loading buffer and gel tracking dyes. Use of CoralLoad PCR Buffer is not recommended without an intermediate purification of the PCR product if downstream applications require fluorescence or absorbance measurements.
When using Q-Solution for the first time in a particular primer–template system, parallel reactions with and without Q-Solution should always be performed.
TA-cloning with PCR products generated using Taq DNA Polymerase
TA-cloning subcloning avoids the use of restriction enzymes and relies on adenine (A) and thymine (T) on different DNA fragments to hybridize and become ligated in the presence of ligase. Taq DNA Polymerase preferentially adds an adenine to the 3'-end of PCR products. These PCR amplified inserts can be cloned into linearized vectors that have complementary 3' thymine overhangs.
Applications
Taq DNA Polymerase is used for standard and specialized applications, including:
- Routine PCR
- RT-PCR
- PCR cloning
- Screening
- STR fingerprinting
- VNTR fingerprinting for the diagnosis of microbial infections
- RAPD PCR fingerprinting
- Differential display
Supporting data and figures
Lot-to-lot reproducibility.
A fragment of the single-copy gene for cystic fibrosis was amplified from 30 ng, 3 ng, and 300 pg human genomic DNA corresponding to 104, 103, and 102 copies of target template, respectively. Three different lots of QIAGEN Taq DNA Polymerase were used and equal volumes of the PCR product were analyzed on a 1% agarose gel. M: markers.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, RT-PCR, DNA fingerprinting, TA cloning |
| dNTP's included | No/Yes (Taq PCR Core Kit) |
| Real-time or endpoint | Endpoint |
| Reaction type | PCR amplification |
| Single or multiplex | Single |
| With/without hotstart | Without hot start |
| Enzyme activity | DNA polymerase with 5’→3’ exonuclease |
| Mastermix | No |
| Sample/target type | Genomic DNA and cDNA |