✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 56304
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Rapid purification of high-quality DNA
- No organic extraction or alcohol precipitation
- Consistent, high yields
- Complete removal of contaminants and inhibitors
- Automated purification of 1–12 samples on the QIAcube Connect
Product Details
QIAamp DNA Kits provide silica-membrane-based nucleic acid purification from tissues, swabs, CSF, blood, body fluids or washed cells from urine. In addition, genomic and mitochondrial DNA can be purified from small amounts of fresh or frozen blood, tissue and dried blood spots. Mechanical homogenization is not required as the tissues are lysed enzymatically. DNA purification from 1–12 samples can be automated on the QIAcube Connect using the dedicated QIAamp DNA Mini QIAcube Kit. Purification of DNA using the QIAamp DNA Micro Kit and QIAamp DNA Mini Kit is also automatable on the QIAcube Connect.
QIAamp DNA Mini standard protocols can also be executed using the TRACKMAN Connected system, paired with PIPETMAN M Connected pipettes, both from Gilson. The TRACKMAN Connected system guides researchers through the QIAamp DNA Mini protocols while automatically adjusting the Bluetooth-enabled PIPETMAN M Connected pipette settings. Each step of the protocol execution is recorded to accelerate reporting by generating a comprehensive run report. Download more information.
Performance
DNA purified using the QIAamp DNA Micro Kit is free of proteins, nucleases and other impurities, and is suitable for use in sensitive downstream applications, such as real-time PCR (see figure " Efficient purification of DNA from small sample sizes") and laser microdissection (LMD) PCR (see figure " Laser microdissection PCR"). Purified DNA may also be used in short-tandem repeat (STR) genotyping, single-nucleotide polymorphism (SNP) genotyping or pharmacogenomic research.
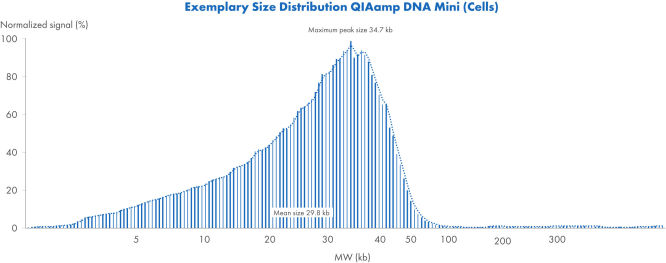
The QIAamp DNA Mini Kit yields DNA sized up to 50 kb (see figure " Purification of up to 50 kb genomic DNA"). DNA of this length denatures completely and has the highest amplification efficiency. Yields of nucleic acids or DNA depend on the starting material (see table “Typical yields with the QIAamp DNA Mini Kit”). DNA purified using the QIAamp DNA Mini Kit can be used in a wide range of downstream applications, including PCR and quantitative real-time PCR, Southern blotting, SNP and STR genotyping and pharmacogenomic research.
| Sample | Amount | Total nucleic acid yields (µg)* | DNA yields (µg)† |
|---|---|---|---|
| Blood | 200 µl | 4–12 | 4–12 |
| Buffy coat | 200 µl | 25–50 | 25–50 |
| Cells | 106 | 20–30 | 15–20 |
| Liver | 25 mg | 60–115 | 10–30 |
| Brain | 25 mg | 35–60 | 15–30 |
| Lung | 25 mg | 25–45 | 5–10 |
| Heart | 25 mg | 15–40 | 5–10 |
| Kidney | 25 mg | 40–85 | 15–30 |
| Spleen | 10 mg | 25–45 | 5–30 |
The QIAamp DNA Mini QIAcube Kit yields DNA sized up to 50 kb: DNA of this length denatures completely and has the highest amplification efficiency. The purified DNA may be used in a number of applications, including:
- Viral research
- Bacterial research
- Fungal research
- Cancer research
- Human genetic testing research
- Paternity testing
- Forensic analysis
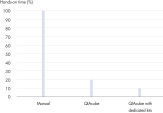
The QIAamp DNA Mini QIAcube Kit simplifies DNA isolation from tissue samples with fast spin-column procedures. The kit includes rotor adapters that are preloaded with QIAamp spin columns and elution tubes, delivering greater convenience and time savings (see figure " Significant time savings").
See figures
Principle
The QIAamp DNA Micro Kit combines the selective binding properties of a silica-based membrane with flexible elution volumes of between 20 and 100 µl. QIAamp DNA Micro technology yields genomic and mitochondrial DNA from small samples ready to use in PCR and blotting procedures.
The QIAamp DNA Mini Kit simplifies DNA isolation from tissue samples with fast spin-column or vacuum procedures. QIAamp DNA technology yields genomic, mitochondrial, bacterial, parasite or viral DNA from human tissue samples ready to use in PCR and blotting procedures.
During the QIAamp DNA purification procedure, DNA binds specifically to the QIAamp MinElute or QIAamp silica-gel membrane while contaminants pass through. No phenol-chloroform extraction is required. PCR inhibitors, such as divalent cations and proteins, are completely removed in two efficient wash steps, leaving pure DNA to be eluted in either water or a buffer provided with the kit. QIAamp DNA technology yields genomic, mitochondrial, bacterial, parasite or viral DNA from human tissue samples ready to use in PCR and blotting procedures.
QIAamp sample preparation technology is fully licensed, allowing QIAamp purified nucleic acids to be used in any molecular assay or other downstream application without risk of patent infringement.
Procedure
The QIAamp DNA Micro Kit simplifies DNA isolation from small samples with a fast spin-column procedure (see flowchart " QIAamp DNA Micro procedure"). The QIAamp DNA Micro Kit uses well-established technology for purification of genomic and mitochondrial DNA from small sample volumes or sizes (e.g., 1–100 µl whole blood, blood cards of ~3 mm diameter, urine or <10 mg tissue or laser microdissected tissue). Fresh or frozen blood, tissue and dried blood spots can be purified in 30 minutes and eluted in 20–100 µl. The QIAamp DNA Micro Kit can also be automated on the QIAcube Connect.
Optimized buffers and enzymes in the QIAamp DNA Mini Kit lyse samples, stabilize nucleic acids and enhance selective DNA adsorption to the QIAamp membrane. Alcohol is added and lysates loaded onto the QIAamp spin column. Wash buffers are used to remove impurities and pure, ready-to-use DNA is then eluted in water or low-salt buffer.
No mechanical homogenization is necessary as the tissues are lysed enzymatically. The convenient spin-column procedure means that hands-on preparation time is only 20 minutes (lysis times differ according to the sample source). Samples can be processed using either a microcentrifuge or, if blood or other body fluids are being processed, using the QIAvac 24 Plus. In addition, the rigorous lysis procedure employed makes the QIAamp DNA Mini Kit ideal for purification of genomic DNA from bacteria or parasites. To further reduce hands-on time, genomic DNA purification may be automated on the QIAcube. The QIAamp DNA Accessory Set A provides the additional buffers and reagents needed for isolation of genomic, mitochondrial, bacterial, parasite or viral DNA using the QIAcube with 12 x QIAamp DNA Mini Kits (50).
Vacuum processing
Blood or other body fluids can be processed by vacuum, instead of centrifugation, for greater speed and convenience in DNA purification. QIAamp Mini spin columns are accommodated on the QIAvac 24 Plus manifold using VacValves and VacConnectors. VacValves should be used if sample flow rates differ significantly, to ensure consistent vacuum. Disposable VacConnectors are used to avoid any cross-contamination. Use of VacConnectors also allows these QIAamp spin procedures to be performed on QIAvac 6S with QIAvac Luer Adapters.
Automated DNA isolation on the QIAcube Connect
The dedicated QIAamp DNA Mini QIAcube Kit is designed with preloaded spin columns and elution tubes in rotor adapters for QIAcube instruments, eliminating the risk of errors due to incorrect loading of rotor adapters. The dedicated kit is tailored to QIAcube requirements, reducing waste.
A simple protocol (lyse, bind, wash and elute) and an automated protocol on the QIAcube Connect requires minimal user interaction. The award-winning QIAcube Connect uses advanced technology to process QIAGEN spin columns, enabling seamless integration of automated, low-throughput sample prep into laboratory workflows. All steps in the purification procedure are fully automated — up to 12 samples can be processed per run. The QIAcube Connect and the dedicated QIAamp DNA Mini QIAcube Kit provide a winning combination for fast, easy and convenient DNA purification.
See figures
Applications
The QIAamp DNA Micro Kit procedure is suitable for a wide range of sample materials, including small volumes of blood, blood cards, urine, small tissue samples, including laser microdissections.
The QIAamp DNA Mini Kit is ideal for purification of DNA from most commonly used human tissue samples, including muscle, liver, heart, brain, bone marrow and other tissues, swabs (buccal, eye, nasal, pharyngeal and others), CSF, blood, body fluids and washed cells from urine. DNA can be purified from up to 25 mg tissue or from up to 200 µl fluid in 20 minutes, and eluted in 50–200 µl.
The QIAamp DNA Mini QIAcube Kit is for automated isolation of genomic, mitochondrial, parasite or viral DNA on the QIAcube Connect. Sample sources include tissues, buccal swabs, CSF and washed cells from urine.
| Features | QIAamp DNA Micro Kit | QIAamp DNA Mini Kit |
|---|---|---|
| Applications | Real-time PCR, STR analysis, LMD-PCR | PCR, Southern blotting |
| Elution volume | 20–100 µl | 50–200 µl |
| Format | Spin column | Spin column |
| Main sample type | Whole blood | Whole blood, tissue, cells |
| Processing | Manual (centrifugation or vacuum) | Manual (centrifugation or vacuum) |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein |
Genomic DNA, mitochondrial DNA | Genomic DNA, mitochondrial DNA, bacterial DNA, parasite DNA, viral DNA |
| Sample amount | 1–100 µl | 200 µl/25 mg/5 x 106 |
| Technology | Silica technology | Silica technology |
| Time per run or per prep | 30 minutes | 20 minutes |
| Yield | <3 µg | 4–30 µg |
Supporting data and figures
Exemplary size distribution using QIAamp DNA Mini Kit (Cells)
Exemplary size distribution of DNA from cells using the QIAamp DNA Mini Kit. Samples were analyzed with the Femto Pulse System and signals were normalized to maximum peak.