✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
QIAamp DNA Micro Kit(50)
カタログ番号 / ID. 56304
✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
特徴
- 高品質DNAの迅速な精製
- 有機溶媒による抽出もアルコールの沈殿も不要
- 一貫性があり、高収量
- 汚染物質や阻害物質の完全な除去
- QIAcube Connectを使った1~12サンプルの自動精製
製品詳細
QIAamp DNA Kitは、組織、スワブ、CSF、血液、体液、尿由来の洗浄細胞からシリカ膜ベースの核酸精製を可能にします。また、少量の新鮮または冷凍された血液、組織、乾燥した血痕からゲノムDNAおよびミトコンドリアDNAを精製できます。組織は酵素で溶解されるため、機械的な均質化は必要ありません。QIAcube Connectと専用のQIAamp DNA Mini QIAcube Kitを使用すれば、1~12サンプルのDNA精製を自動化できます。また、QIAamp DNA Micro KitおよびQIAamp DNA Mini Kitを使用したDNA精製もQIAcube Connectで自動化することが可能です。
QIAamp DNA Mini標準プロトコールは、TRACKMAN Connected systemを使って実行することもできます。
パフォーマンス
QIAamp DNA Micro Kitは、シリカベース膜の選択的結合特性と20~100 µLに対応可能な柔軟な溶出量とを兼ね備えています。QIAamp DNA Micro Kitを使って精製したDNAには、タンパク質、ヌクレアーゼなどの不純物が含まれておらず、real-time PCR(図「 少量のサンプルからのDNAの効果的な精製」を参照)やレーザー顕微解剖(LMD)PCR(「 レーザー顕微解剖PCR」を参照)のような高感度のダウンストリームアプリケーションに適しています。精製DNAは、ショートタンデムリピート(STR)ジェノタイピング、一塩基多型(SNP)ジェノタイピング、またはファーマコゲノミクスにも使用できます。
QIAamp DNA Mini Kitは、高速スピンカラム法または真空プロトコールを用いることで、組織からのDNA分離を簡略化し、最大50 kbの長さのDNAを回収します(「 最大50 kbのゲノムDNAの精製」を参照)。この長さのDNAは完全に変性し、最も高い増幅効率を示します。核酸またはDNAの収量は、使用する出発物質に依存します(表「QIAamp DNA Mini Kitの標準的収量」を参照)。QIAamp DNA Mini Kitを用いて精製したDNAは、PCR、定量real-time PCR、サザンブロッティング、SNPおよびSTRジェノタイピング、ファーマコゲノミクス研究など、さまざまなダウンストリームアプリケーションに使用できます。
| サンプル | 量 | 核酸 収量の合計(µg)* | DNA収量 (µg)† |
|---|---|---|---|
| 血液 | 200 µL | 4~12 | 4~12 |
| 軟膜 | 200 µL | 25~50 | 25~50 |
| 細胞 | 106 | 20~30 | 15~20 |
| 肝臓 | 25 mg | 60~115 | 10~30 |
| 脳 | 25 mg | 35~60 | 15~30 |
| 肺 | 25 mg | 25~45 | 5~10 |
| 心臓 | 25 mg | 15~40 | 5~10 |
| 腎臓 | 25 mg | 40~85 | 15~30 |
| 脾臓 | 10 mg | 25~45 | 5~30 |
QIAamp DNA Mini QIAcube Kitを使用することで、最大50 kbのDNAを回収できます。この長さのDNAは完全に変性し、最も高い増幅効率を示します。精製されたDNAは、以下のようなさまざまなアプリケーションに使用できます
- ウイルス研究
- 細菌研究
- 菌研究
- がん研究
- ヒト遺伝子検査研究
- 父子鑑定
- 法医学的分析
QIAamp DNA Mini QIAcube Kitは、高速スピンカラム法を用いることで組織からのDNA分離を簡略化します。キットには、あらかじめQIAampスピンカラムと溶出チューブが搭載されたローターアダプターが含まれており、時間と手間を大幅に節約できます(図「 時間と手間を大幅に節約」を参照)。
原理
QIAamp DNA精製プロセスでは、DNAはQIAamp MinEluteまたはQIAampシリカゲル膜に特異的に結合し、汚染物質は通過します。フェノール–クロロホルム抽出は必要ありません。2価カチオンやタンパク質などのPCR阻害物質は、2回の効率的な洗浄工程で完全に除去され、純粋なDNAのみが残り、水またはキット付属のバッファーに溶出されます。
QIAamp DNA技術は、ヒト組織サンプルから、PCRやブロッティングにすぐに使用できるゲノムDNA、ミトコンドリアDNA、細菌DNA、寄生虫DNA、ウイルスDNAの抽出を可能にします。
QIAampサンプル調製技術は正式にライセンスを取得しており、QIAampで精製された核酸は、特許侵害の懸念なく、あらゆる分子アッセイやダウンストリームアプリケーションに使用できます。
操作手順
最適化されたバッファーとQIAamp DNA Mini Kitの酵素でサンプルを溶解し、核酸を安定化させ、QIAamp膜に対する選択的DNA吸着を強化します。アルコールを加え、溶解物をQIAampスピンカラムにロードします。
洗浄バッファーを使用して不純物を除去すると、高純度ですぐに使えるDNAが水または低塩バッファーに溶出します。
組織は酵素的に溶解されるため、機械的均質化は必要ありません。便利なスピンカラムを使用したプロセスにより、実際の調製時間はわずか20分で済みます(溶解時間はサンプルの種類によって異なります)。
サンプルはマイクロ遠心機、または血液やその他の体液を処理する場合はQIAvac 24 Plusを使用して処理できます。また、精密な溶解プロセスを採用しているため、QIAamp DNA Mini Kitは細菌や寄生虫からのゲノムDNA精製に最適です。
ゲノムDNA精製をQIAcubeで自動化することで、実際の操作時間をさらに短縮することができます。QIAamp DNA Accessory Set Aには、QIAcubeと12 x QIAamp DNA Mini Kit(50)を使用して、ゲノムDNA、ミトコンドリアDNA、細菌DNA、寄生虫DNA、ウイルスDNAを分離するために必要な追加のバッファーと試薬が含まれています。
真空プロトコール
血液やその他の体液は、遠心分離ではなく真空法で処理することができ、これによりDNA精製の速度と利便性が大幅に向上します。QIAamp Miniスピンカラムは、QIAvac 24 PlusマニホールドにVacValveおよびVacConnectorを使用して設置されています。
サンプルの流量が著しく異なる場合は、一貫した真空状態を確保するためにVacValvesを使用する必要があります。使い捨てのVacConnectorsは、クロスコンタミネーションを防止するために使用されます。さらに、VacConnectorsを使用することで、QIAvac 6SにQIAvac Luer Adaptersを取り付けてQIAampスピン処理を実施することも可能です。
QIAcube ConnectによるDNA分離の自動化
専用のQIAamp DNA Mini QIAcube Kitは、QIAcube機器で使用するために、あらかじめローターアダプターにスピンカラムと溶出チューブを搭載しており、ローターアダプターの誤搭載によるエラーのリスクを防ぎます。この専用キットはQIAcubeの要件に合わせて設計されており、廃棄物の削減にも寄与します。
QIAcube Connectにはシンプルなプロトコール(溶解、結合、洗浄、溶出)と自動化されたプロトコールが搭載されており、ユーザーが行う操作は最小限に抑えられています。
アワードを受賞したQIAcube Connectは、QIAGENスピンカラムの処理に最新技術を活用し、自動化された低スループットのサンプル調製をラボのワークフローにシームレスに組み込むことができます。精製プロセスのすべての工程は完全に自動化され、1回のランで最大12サンプルを処理できます。QIAcube Connectと専用のQIAamp DNA Mini QIAcube Kitを組み合わせることで、高速で簡単、かつ便利なDNA精製が可能となります。
アプリケーション
QIAamp DNA Micro Kitは、少量の血液、乾燥血液スポットカード、尿、レーザーマイクロダイセクションによる切片といった小さい組織サンプルなど、広範なサンプルの処理に適しています。
QIAamp DNA Mini Kitは、筋肉、肝臓、心臓、脳、骨髄などのよく使用されるヒト組織やスワブ(頬粘膜、眼、鼻腔、咽頭、その他)、CSF、血液、体液、尿由来の洗浄細胞からのDNA精製に最適です。組織は最大25 mg、液体は最大200 µLを20分で精製し、50~200 µLを溶出可能です。
QIAamp DNA Mini QIAcube Kitは、QIAcube Connectを使用して、ゲノムDNA、ミトコンドリアDNA、寄生虫DNA、ウイルスDNAの自動分離を行うためのキットです。サンプルの由来には、組織、頬粘膜スワブ、CSF、尿由来の洗浄細胞などがあります。
| 特徴 | QIAamp DNA Micro Kit | QIAamp DNA Mini Kit |
|---|---|---|
| アプリケーション | Real-time PCR、STR解析、LMD-PCR | PCR、サザンブロッティング |
| 溶出量 | 20~100 µL | 50~200 µL |
| フォーマット | スピンカラム | スピンカラム |
| 主なサンプルタイプ | 全血 | 全血、組織、細胞 |
| 処理 | 手動(遠心分離または真空分離) | 手動(遠心分離または真空分離) |
| トータルRNA、miRNA、 poly A+ mRNA、DNA、タンパク質の精製 | ゲノムDNA、ミトコンドリアDNA | ゲノムDNA、ミトコンドリアDNA、 細菌DNA、寄生生物DNA、ウイルスDNA |
| サンプル量 | 1~100 µL | 200 µL/25 mg/5 x 106 |
| 技術 | シリカテクノロジー | シリカテクノロジー |
| ランあたり、または調製あたりの時間 | 30分 | 20分 |
| 収量 | 3 µg未満 | 4~30 µg |
裏付けデータと数値
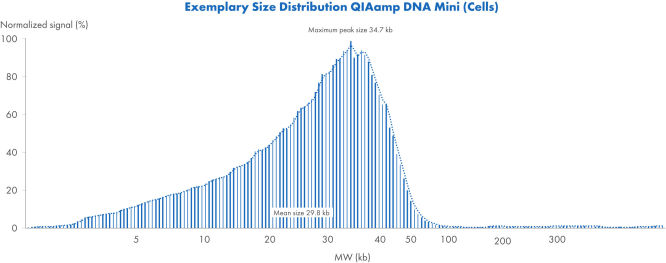
QIAamp DNA Mini Kit (細胞)を使ったときの典型的な大きさの分布
QIAamp DNA Mini Kitを使用して細胞から得られたDNAの典型的なサイズ分布です。サンプルはFemto Pulse Systemで分析され、信号は最大ピークに基づいて正規化されました。