QuantiTect Reverse Transcription Kit
快速合成cDNA,进行两步法real-time RT-PCR,用于基因表达分析
快速合成cDNA,进行两步法real-time RT-PCR,用于基因表达分析
目录编号 / ID. 205311
QuantiTect Reverse Transcription Kit通过方便快速的操作过程进行cDNA合成,并有效去除基因组DNA。应用gDNA Wipeout Buffer可高效去除RNA样本中的基因组DNA污染。QuantiTect Reverse Transcription Kit提供所有快速、高效逆转录所需的组分,包括Quantiscript Reverse Transcriptase、Quantiscript RT Buffer和独特的RT Primer Mix。合成的cDNA经过优化可用于real-time PCR,实现对mRNA转录本所有区域的靶分子进行可靠的定量检测。
使用QuantiTect Reverse Transcription Kit,基因组DNA污染物可使用独特的gDNA Wipeout Buffer迅速有效的去除(参见" Effective genomic DNA removal for accurate real-time RT-PCR")。去除基因组DNA对于准确的基因表达结果非常重要,而并不总是可以设计RNA特异性引物或探针。使用gDNA Wipeout Buffer可节省时间和费用,因为纯化RNA样品过程中或纯化后均无需另外采用DNase消化。
Quantiscript Reverse Transcriptase的高RNA亲和性配合Quantiscript RT Buffer可从各种RNA模板中获得高产量cDNA(见表“低丰度转录本可获得较高产量的cDNA”)。即使复杂的模板也能成功逆转录,如GC含量高或有复杂二级结构的模板。
| IL12A的CT值 (低表达) | IL1RN的CT值 (高表达) | |||
| Input RNA (ng) | QIAGEN | Supplier AII | QIAGEN | Supplier AII |
|---|---|---|---|---|
| 1000 | 30.9 | 32.0 | 23.1 | 24.9 |
| 100 | 34.2 | 35.4 | 26.3 | 26.6 |
| 10 | 37.8 | 46.8 | 29.7 | 30.3 |
| 1 | N.D. | N.D. | 32.4 | 34.5 |
RT Primer Mix包含有特别优化的oligo-dT和随机引物的混合物,使cDNA合成可从RNA转录本的各个区域起始,甚至可从5'区域开始(参见" Sensitive detection of a target at the 5' region of a 12.5 kb transcript")。与其他供应商提供的试剂盒相比,QuantiTect Reverse Transcription Kit为real-time PCR分析提供高产量的cDNA,无需考虑待扩增目标序列位于转录本的哪个区域,检测低丰度的基因也具有更高灵敏度(参见" Higher sensitivity in real-time, two-step RT-PCR")。同时,QuantiTect Reverse Transcription Kit也确保了real-time RT-PCR结果更好的重复性。
QuantiTect Reverse Transcriptase是Omniscript和Sensiscript Reverse Transcriptases的新型混合物,对RNA具有高亲和性,可从各种含量的RNA(10 pg到1 µg)合成cDNA。与其他供应商提供的试剂盒相比,QuantiTect Reverse Transcription Kit为real-time PCR分析提供高产量的cDNA,无需考虑待扩增目标序列位于转录本的哪个区域。即使复杂的模板也能成功逆转录,如GC含量高或有复杂二级结构的模板。QuantiTect RT Buffer已经过优化,可与real-time PCR缓冲液兼容。
为获得准确的real-time RT-PCR基因表达分析结果,很重要的是仅扩增和检测cDNA。基因组DNA的干扰可通过设计横跨外显子/外显子边界的引物或探针避免。尽管如此,在某些特殊情况下(如:cDNA来自一个单外显子基因),起始RNA样品必须无基因组DNA。使用QuantiTect Reverse Transcription Kit,基因组DNA污染物可使用独特的gDNA Wipeout Buffer迅速有效的去除。使用gDNA Wipeout Buffer可节省时间和费用,因为纯化RNA样品过程中或纯化后均无需另外采用DNase消化。另外,不需要设计RNA特异性引物或探针。
| 组份 | 优势 |
|---|---|
| gDNA Wipeout Buffer | 在real-time RT-PCR时仅检测RNA |
| Quantiscript Reverse Transcriptase | 适用的RNA起始量范围大(10 pg到1 µg RNA) 灵敏度高 |
| Quantiscript RT Buffer | 处理复杂模板 |
| RT Primer Mix | 对转录本的各个区域,包括5'区进行cDNA合成 |
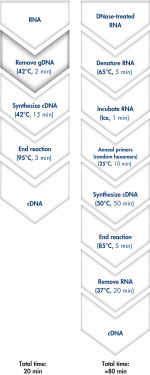
使用QuantiTect Reverse Transcription Kit可在20分钟内去除基因组DNA和合成cDNA(参见" Fast and convenient cDNA synthesis")。由于两个反应均在同一孵育温度并使用预混液构建反应,因此操作过程快速且方便。
QuantiTect Reverse Transcription Kit包括快速合成cDNA所需的所有组分。纯化的RNA只需简单地与gDNA Wipeout Buffer孵育,即可有效去除污染的基因组DNA。与其他的方法相比,使用Quantiscript Reverse Transcriptase、Quantiscript RT Buffer和RT Primer Mix制备的预混液可将RNA样品直接进行逆转录。使用Quantiscript Reverse Transcriptase,即使具有复杂的二级结构的RNA也可在低温下转录,确保RNA保持完整:整个反应都在42°C下完成,并在95°C失活。不需要RNA变性、引物退火和RNase H消化。
QuantiTect Reverse Transcription Kit可从各种类型的起始样本中合成cDNA,以实现高效和高灵敏度的real-time RT-PCR。样本类型包括激光显微切割样本和活组织切片。
Genomic DNA removal and cDNA synthesis take only 20 minutes with the QuantiTect Reverse Transcription Kit. The procedure is fast and convenient since both reactions are run using the same incubation temperature and are set up using master mixes. In contrast, the procedure for the kit from Supplier I is much longer and requires more "hands-on time" due to additional pipetting steps and frequent changes in incubation temperature.
| 特点 | 规格 |
|---|---|
| Applications | Quantification of (even low-abundance) transcripts |
| Sample/target type | RNA template |
| Enzyme activity | Reverse transcription |
| Real-time or endpoint | Real time |
| Reaction type | Two-step, cDNA production, genomic DNA digestion |
| Single or multiplex | Single |
| Mastermix | No |