✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
DNeasy UltraClean 96 Microbial Kit (384)
Cat. No. / ID: 10196-4
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Fast and easy DNA and protein purification with spin filter extraction
- High-throughput DNA extraction in 96-well format
- Efficient lysis with bead-beating protocol
- Extraction of total cellular proteins, plus microbial protein from soil
- Ready-to-use, high-quality DNA and proteins for downstream applications
Product Details
DNeasy UltraClean/NoviPure Microbial Kits enable extraction of high-quality genomic DNA from a variety of microorganisms, including yeast, bacteria and fungi. The kits also allow for extraction of total cellular protein from fungi, Gram-positive and Gram-negative bacteria and other microbial cultures. Extracellular and intracellular microbial protein can be isolated from up to 5 g of any type of soil, without co-extraction of interfering compounds, such as humic substances.
Request a quote for a trial DNeasy UltraClean Microbial Kit.
Isolation of DNA using the DNeasy UltraClean Microbial Kit can be automated on the QIAcube Connect.
Performance
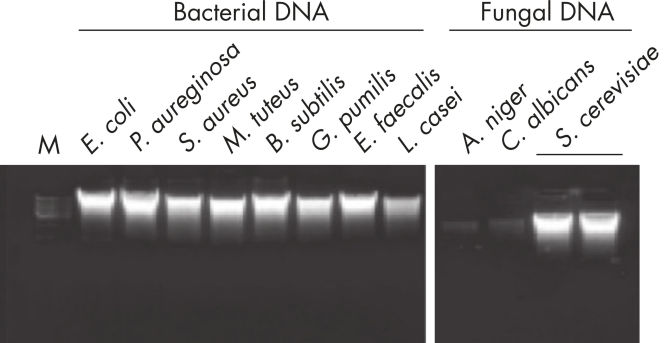
The DNeasy UltraClean Microbial procedure extracts high-quality DNA from a range of microorganisms (see figure “DNeasy UltraClean purifies high-quality DNA from a range of microorganisms”).
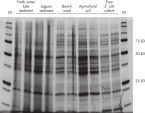
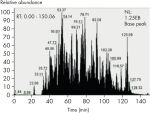
Protein purification using NoviPure procedures enables extraction of high yields of pure protein (see figures “High yields of protein from all soil types with the NoviPure Soil Protein Kit” and “Pure protein extraction from soil with the NoviPure Soil Protein Kit”). Purified protein gave high-quality results in demanding downstream applications (see figure “Improved protein results with the NoviPure Soil Protein Kit”).
See figures
Procedure
The DNeasy UltraClean Microbial procedure uses a bead-beating protocol to extract DNA. Microbial cells are resuspended in a bead solution and added to a bead beating tube or plate containing beads. Then, lysis solution is added. The microorganisms are lysed by a combination of heat, detergent and mechanical force due to the impact force and shear effect of the specialized beads. The cellular components are lysed by mechanical action using a specially designed Vortex Adapter on a standard vortex or 96-well plate shaker. The DNA released from the lysed cells is bound to a silica spin filter or spin plate. The spin filter is washed, and the DNA is recovered in DNA-free Tris buffer.
DNA purification from up to 12 samples can be automated on the QIAcube Connect.
The NoviPure procedure uses a bead-beating protocol in the presence of a strong chaotropic agent to efficiently lyse and solubilize total proteins from a large variety of microbial species, including fungi, Gram-negative and Gram-positive bacteria. The use of silica spin filters (in the MB Spin Columns) to achieve reversible immobilization of proteins – a patent-pending technical advance in the chemistry of protein extraction – greatly simplifies protein isolation by removing tedious and bias-inducing protein precipitation steps. Following in-gel trypsin digestion, isolated proteins are suitable for downstream applications such as 1D SDS-PAGE and mass spectrometry.
Denatured microbial protein can be extracted from a wide range of soil types using a two-buffer bead-beating protocol to efficiently lyse cells while solubilizing intracellular as well as extracellular protein. The protein is precipitated, and the pellet is washed to remove residual salts and detergent. The protein pellet can be resuspended in any buffer for further analysis or storage.
Applications
Isolated nucleic acids (gDNA, rRNA, mRNA and small RNAs) are suitable for demanding downstream applications, including PCR, qPCR, RT-PCR and next-generation sequencing (NGS).
Extracted proteins can be used in a wide range of downstream applications, including mass spectrometry and 1D or 2D polyacrylamide gel electrophoresis.
Comparison of DNeasy UltraClean/NoviPure Microbial Kits
| Features | DNeasy UltraClean 96 Microbial Kit | NoviPure Microbial Protein Kit | NoviPure Soil Protein Kit |
|---|---|---|---|
| Bead size | 0.1 mm glass | 0.1 mm glass, 0.5 mm glass mix | 0.1 mm glass, 0.1 mm ceramic mix |
| Binding capacity | Up to 20 µg per prep | – | – |
| Format | Silica Spin Filter Plates | Silica Spin Filter | Patent-pending two buffer extraction method |
| Processing | Bead beating | Bead beating | Bead beating |
| Sample size | 1.8 ml | 1.8 ml of microbial culture; QIAGEN recommends using no more than 1x108 fungal cells or 1x109 bacterial cells per sample | 2–5 grams |
| Sample types | Processed cultured Gram-negative bacteria and Gram-positive bacteria, yeast, fungi | Processed cultured fungi, Gram-negative and Gram-positive bacteria | Processed all types of soils, sand, and sediment |
| Storage temperature | Store at room temperature (15–30°C) | Store spin filters at 4°C. All other kit reagents and components can be stored at room temperature (15-30°C) | Store at room temperature (15-30°C) Pre-chill Solution SP1 and Solution SP2 before use |
| Throughput | 96 samples | 1–24 samples | 1–2 samples |
| Time per run or per prep | <1.5 hours | 22 minutes | – |
Supporting data and figures
DNeasy UltraClean purifies high-quality DNA from a range of microorganisms.
Results shown on 1.2% TAE agarose gel. Lower cell densities in starting cultures resulted in lower DNA yields in A. niger and C. albicans lanes.