High PCR specificity를 위한 Premixed solution
HotStarTaq Master Mix Kit (250 U)
Cat. No. / ID: 203443
특징
- Hot start tag 사용으로 non-specific amplification을 줄임
- 쉬운 사용방법 — 편리한 master mix로상온에서 간단하게 reaction setup이 가능함
- Optimization 최소화 — 최소의 시간과 비용
제품 세부 정보
HotStarTaq Master Mix contains HotStarTaq DNA Polymerase, the unique QIAGEN PCR Buffer that minimizes the requirement for optimization, and dNTPs. Providing all components in a master mix reduces pipetting steps and the risk of contamination, while increasing throughput and reproducibility.
성능
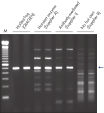
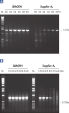
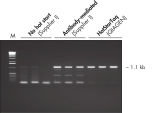
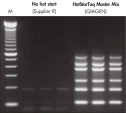
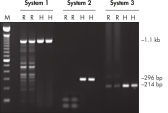
Each lot of HotStarTaq Master Mix Kit is subjected to a comprehensive range of quality control tests, including a stringent PCR specificity and reproducibility assay in which low-copy targets are amplified. HotStarTaq Master Mix Kit outperformed kits tested from other suppliers and ensures high specificity and superior performance in hot-start PCR (see figures "Higher specificity with different primer–template systems" and " Superior performance " and table). The innovative PCR buffer provided with the kit ensures specificity over a wide range of PCR conditions, minimizing the need for optimization (see figures " Wide annealing temperature window" and " Tolerance to variable magnesium concentration").
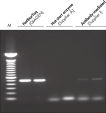
The combination of high specificity and easy handling makes the HotStarTaq Master Mix Kit suitable for use with complex genomic or cDNA templates (see figure " Effect of hot start on RT-PCR performance"), multiple primer pairs (see figure " Specific amplification in multiplex PCR"), and templates isolated from difficult sources or very low-copy targets (see figure " Highly sensitive single-cell PCR"). It is also suitable for projects such as genetic screening, in which large numbers of samples are amplified.
| HotStarTaq DNA Polymerase | Hot-start enzyme from Supplier AII | Antibody-mediated | Manual | Wax barrier | |
|---|---|---|---|---|---|
| Specific amplification | ++ | + | + | +/– | +/– |
| Minimal PCR optimization | ++ | +/– | +/– | – | – |
| Easy to use | ++ | ++ | + | – | – |
HotStarTaq DNA Polymerase specifications
Concentration: 5 units/µl
Recombinant enzyme: Yes
Substrate analogs: dNTP, ddNTP, dUTP, biotin-11-dUTP, DIG-11-dUTP, fluorescent-dNTP/ddNTP
Extension rate: 2–4 kb/min at 72°C
Half-life: 10 min at 97°C ; 60 min at 94°C
Amplification efficiency: ≥105 fold
5'–>3' exonuclease activity: Yes
Extra A addition: Yes
3'–>5' exonuclease activity: No
Contaminating nucleases: No
Contaminating RNases: No
Contaminating proteases: No
Self-priming activity: No
그림 참조
원리
HotStarTaq Master Mix is a ready-to-use mixture of HotStarTaq DNA Polymerase, QIAGEN PCR Buffer, and dNTPs. HotStarTaq DNA Polymerase, a modified form of Taq DNA Polymerase, provides high specificity in hot-start PCR.
HotStarTaq DNA Polymerase
HotStarTaq DNA Polymerase is supplied in an inactive state and has no polymerase activity at ambient temperatures. This prevents extension of nonspecifically annealed primers and primer dimers formed at low temperatures during PCR setup and the initial PCR cycle (see figures " Superior performance in hot-start PCR" and "Higher specificity with different primer–template systems"). HotStarTaq DNA Polymerase is activated by a 15-minute incubation at 95°C, which can be incorporated into any existing thermal-cycler program.
QIAGEN PCR Buffer
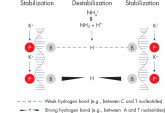
QIAGEN PCR Buffer maintains specific amplification in every cycle of PCR by promoting a high ratio of specific-to-nonspecific primer binding during the annealing step in each PCR cycle (see figure " Increased specificity of primer annealing"). Owing to a uniquely balanced combination of KCl and (NH4)2SO4, the buffer provides stringent primer-annealing conditions over a wider range of annealing temperatures and Mg2+ concentrations than conventional PCR buffers. Optimization of PCR by varying the annealing temperature or the Mg2+ concentration is therefore often minimal or not required (see figures " Wide annealing temperature window" and " Tolerance to variable magnesium concentration").
그림 참조
절차
응용 분야
HotStarTaq Master Mix Kit is highly suitable for a wide variety of applications, including challenging applications such as amplification of:
- Complex genomic templates
- Complex cDNA templates (e.g., RT-PCR)
- Very low-copy targets (e.g., single-cell PCR)
- Reactions with multiple primer pairs
지원되는 데이터 및 수치
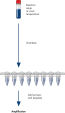
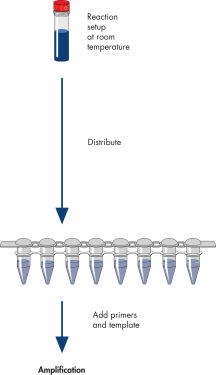
HotStarTaq procedure.
The HotStarTaq procedure is fast and easy for maximum convenience.
Specifications
| Features | Specifications |
|---|---|
| Applications | PCR, RT-PCR, Complex genomic templates, very low-copy targets |
| Real-time or endpoint | Endpoint |
| Mastermix | Yes |
| Enzyme activity | 5'-> 3' exonuclease activity |
| Sample/target type | Genomic DNA and cDNA |
| Single or multiplex | Single |
| Reaction type | PCR amplification |
| With/without hotstart | With hotstart |