✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 250221
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Premixed master mix with controls for easy set up and detection of host cell DNA
- Enables accurate detection of residual host-cell DNA from E.coli, CHO and HEK293 cells down to low femtogram levels
- A multi-copy target assay that detects and quantifies highly fragmented host cell DNA
- Enables host cell DNA detection in both extracted and unextracted samples
Product Details
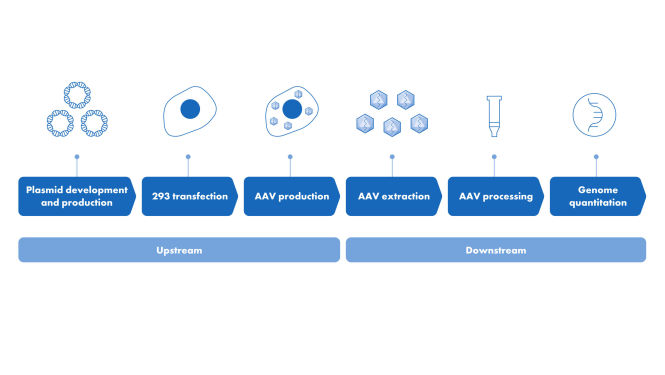
Host cell DNA contamination in products such as vaccines, drugs and other biopharmaceuticals imposes great health risks. Therefore, its safe limits are tightly regulated by agencies such as the U.S. FDA and WHO. Clear guidelines for the upper limits of residual DNA are established based on the nature of drug administration, infectivity and oncogenicity of the contaminating cell DNA. For instance, parenteral administration of non-tumorigenic cell DNA should be limited to 10 ng/dose and a maximum length of 200 bp, whereas less than 100 µg/dose of residual DNA is recommended by WHO for orally administered vaccines.
Detection and removal of such contaminations in manufacturing products require highly sensitive and accurate measurements of the extremely low amounts of specific host cell DNA present in the products.
Digital PCR is the preferred method of detection for residual DNA quantification. This is due to its unmatched sensitivity and accuracy, especially at trace levels of contamination compared to other detection methods such as qPCR. QIAcuity Residual DNA Quantification Kits provide the accurate and precise detection of Escherichia coli (E.coli), Chinese Hamster Ovary cell (CHO) and Human Embryonic Kidney 293 cells (HEK293) host cell DNA.
Performance
The QIAcuity Residual DNA Quantification Kits provide accurate CHO, E.coli and HEK293 residual DNA (resDNA) quantification results with or without extraction, even in the presence of PCR contaminants or other inhibitory reagents. The assays are multicopy target assays that accurately determine highly fragmented residual host cell DNA.
| Assay | Target Copy Number | Amplicon Size | Conversion Factor cp/µl to fg/µl |
|---|---|---|---|
| QIAcuity E.coli resDNA Quant Kit | 7 | <200 | 0.35 |
| QIAcuity CHO resDNA Quant Kit | ~ 1 million, undefined (repeated element) | <100 | 0.28 |
| QIAcuity HEK293 resDNA Quant Kit | ~ 1 million, undefined (repeated element) | <100 | 1.54 |
QIAcuity E.coli resDNA Quant Kit detects as low as 5 fg of residual DNA in a single reaction
| Loading amount per reaction (fg/rxn) | E.coli Standard (copies/µl) | Internal Control (copies/µl)* |
|---|---|---|
| 50000 | 2949.7 | 99.9 |
| 5000 | 291.6 | 91.7 |
| 500 | 27.6 | 91.4 |
| 50 | 2.8 | 93.2 |
| 25 | 1.46 | 92.5 |
| 5 | 0.45 | 90.6 |
| NTC | 0 | 98.9 |
*100 copies/µl expected for Internal Control
Digital PCR provides higher sensitivity of detection at a lower template input range in comparison to qPCR and therefore enables a more robust application.
QIAcuity CHO resDNA Quant Kit detects as low as 5 fg of residual DNA in a single reaction
| Loading amount per reaction (fg/rxn) | CHO Standard (copies/µl) | Internal Control (copies/µl)* |
|---|---|---|
| 50000 | 4939.3 | 108.7 |
| 5000 | 508.3 | 104.9 |
| 500 | 50.2 | 97.6 |
| 50 | 4.7 | 99.5 |
| 25 | 2.6 | 101.3 |
| 5 | 0.6 | 100.4 |
| NTC | 0 | 104.7 |
*100 copies/µl expected for Internal Control
QIAcuity HEK293 resDNA Quant Kit detects as low as 5 fg of residual DNA in a single reaction
| Loading amount per reaction (fg/rxn) | HEK293 Standard (copies/µl) | Internal Control (copies/µl)* |
|---|---|---|
| 50000 | 1104.9 | 96.6 |
| 5000 | 104.5 | 92.5 |
| 500 | 8.83 | 92.4 |
| 50 | 0.99 | 94.4 |
| NTC | 0 | 96.2 |
*100 copies/µl expected for Internal Control
The kits work in conjunction with the QIAcuity Digital PCR System and QIAcuity Nanoplates, offering an end-to-end fast dPCR workflow comparable to qPCR, but delivering absolute quantification of resDNA in your sample. The kits have been designed with the requirements of bioprocess manufacturing and QC in mind.
Principle
The principle of the dPCR reaction in the nanoplates is described here.
Using QIAcuity digital PCR for host cell DNA monitoring improves the LOD/LOQ through partitioning of the bulk sample with the help of QIAcuity Nanoplate 26k. Partitioning increases the effective concentration of the target, allowing a small amount of host cell DNA to be captured and measured with higher accuracy and precision. The ability to load high template input amounts in the QIAcuity Nanoplate 26k combined with an increased number of partitions is one of the reasons for the higher accuracy and precision of residual host cell detection.
Procedure
The lysate is added to the residual quant assay together with the internal control and host cell DNA measured by absolute quantification. Copies/µl is converted to fg/µl via a provided conversion factor.
Applications
The QIAcuity Residual DNA Quantification Kits are ideal for the highly precise quantification of host cell DNA in complex bioprocess intermediates.
Supporting data and figures
Only limited levels of impurities are allowed in final substance
Carryover of host cell DNA poses a safety concern and is strictly regulated by authorities such as FDA and WHO