✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 47014
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Efficient lysis of bacteria and fungi in all soil types
- Up to 8-fold higher DNA yields compared to alternative methods
- Recovery of inhibitor-free DNA for direct use in NGS applications
- Unbiased results accurately represent sample alpha diversity
- Automation on the QIAcube Connect
Product Details
Extracting microbial DNA from soil samples can be challenging. QIAGEN’s new DNeasy PowerSoil Pro Kit is even more effective than our original PowerSoil technology at isolating high yields of pure microbial DNA from all soil types, including compost, clay and top soil. The kit features a novel bead tube and optimized chemistry for more efficient lysis of soil bacteria and fungi. The kit also contains streamlined Inhibitor Removal Technology (IRT) to eliminate the challenging inhibitors commonly found in soil and environmental samples in even less time. Sequencing results reveal higher alpha diversity as measured by observed operational taxonomic units (OTUs) compared to other tested methods. Extraction of DNA using the DNeasy PowerSoil Pro Kit can be automated on the QIAcube Connect.
Want to try this solution for the first time? Request a quote for a trial kit.
Performance
Significantly increased DNA yields from challenging samples
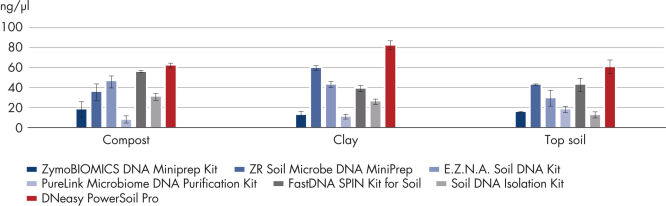
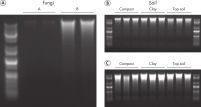
The DNeasy PowerSoil Pro Kit includes a novel bead tube and improved lysis chemistry, which enables isolation of up to 8-fold higher yields of DNA compared to the first generation DNeasy PowerSoil Kit and those of competitors in all soil types tested (see Isolate more high-quality genomic DNA). Both the DNeasy PowerSoil Kit and the new DNeasy PowerSoil Pro Kit isolated high-quality DNA from challenging soil types (see Improved, high-quality DNA yields with the new DNeasy PowerSoil Pro Kit).
Unbiased identification of total diversity and community representation in soil samples
DNA isolated using the DNeasy PowerSoil Pro Kit identified more bacteria (OTUs) in soil samples when compared with the DNeasy PowerSoil Kit and those of two competitors (see Increased bacterial OTUs with the new DNeasy PowerSoil Pro Kit).
See figures
Principle
Inhibitor Removal Technology increases purity of isolated DNA
The DNeasy PowerSoil Pro Kit features a streamlined Inhibitor Removal Technology (IRT) to decrease sample processing time. In a comparison of methods for DNA isolation from soil, the DNeasy PowerSoil Pro Kit was the only method showing 260/280 ratios near 1.8 for all soil types, as well as the highest 260/230 ratios, indicating the absence of inhibitors. Samples isolated using the kit also showed no inhibition in PCR, in contrast to competitor methods (see Increased purity and less variability in DNA extracted with the DNeasy PowerSoil Pro Kit).
See figures
Procedure
Supporting data and figures
Figure 1: Isolate more high-quality genomic DNA.