Configure at GeneGlobe
Find or custom design the right target-specific assays and panels to research your biological targets of interest.
Microbial DNA qPCR Assay
Cat. No. / ID: 330025
One 100 µl tube Microbial DNA qPCR Assay, one 1.35 ml tube Microbial qPCR Mastermix
Log in To see your account pricing.
Microbial DNA qPCR Assays are intended for molecular biology applications. These products are not intended for the diagnosis, prevention, or treatment of a disease.
Configure at GeneGlobe
Find or custom design the right target-specific assays and panels to research your biological targets of interest.
Features
- Experimentally verified assays with high sensitivity and specificity
- Detect microbial species, virulence genes or antibiotic resistance genes
- Simple qPCR workflow
Product Details
Microbial DNA qPCR Assays are a mix of two PCR primers (10 µM each) and one 5′-hydrolysis probe (5 µM) that enables quantitative real-time PCR for 100 reactions. Microbial DNA qPCR Assays are designed using a proprietary and experimentally verified algorithm, providing uniform PCR efficiency and amplification conditions. Each Microbial DNA qPCR Assay undergoes rigorous experimental verification to ensure high PCR efficiency, and this high efficiency is guaranteed when the assays are used with Microbial qPCR Mastermixes and Microbial DNA-Free Water (sold separately).
What is the difference between microbial identification and profiling? Identification is determining the microbe’s presence or absence in your sample which requires you to run a No Template Control during your analysis. Profiling is determining the microbe’s relative expression in two or more experimental conditions and you will need to run a reference sample and a normalizer (provided by QIAGEN).
What is the difference between microbial identification and profiling? Identification is determining the microbe’s presence or absence in your sample which requires you to run a No Template Control during your analysis. Profiling is determining the microbe’s relative expression in two or more experimental conditions and you will need to run a reference sample and a normalizer (provided by QIAGEN).
Performance
Lower limit of quantification (LLOQ)
The LLOQ is the lowest concentration of template that still falls into the linear range of the standard curve (see figure, Limit of detection versus lower limit of quantification). Across all Microbial DNA qPCR Assays, 93% have an LLOQ of <100 gene copies (see figure, The LLOQ for all Microbial DNA qPCR Assays reveals high sensitivity). 92% of microbial identification assays meet this LLOQ, as do 95% of virulence factor gene assays and 97% of antibiotic resistance gene assays (see figures, The LLOQ for microbial identification Microbial DNA qPCR Assays reveals high sensitivity, The LLOQ for virulence factor genes Microbial DNA qPCR Assays reveals high sensitivity, and The LLOQ for antibiotic resistance genes Microbial DNA qPCR Assays reveals high sensitivity).Specificity
Each Microbial DNA qPCR Assay is stringently tested to ensure that it detects only one target species or gene (see figure, Microbial DNA qPCR Assays are highly specific). For assays that do detect more than one target, a list of detected targets and in silico predictions is included on the product sheet.This specificity is maintained even when samples have high species complexity, such as in stool, sputum, and plaque (see figure, Microbial DNA qPCR Assays display high sensitivity even in complex metagenomic samples).
See figures
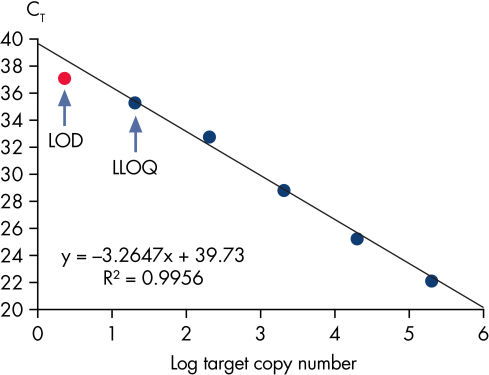
Limit of detection versus lower limit of quantification.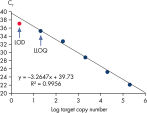 The lower limit of quantification (LLOQ) for all Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for all Microbial DNA qPCR Assays reveals high sensitivity.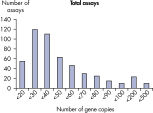 The lower limit of quantification (LLOQ) for microbial identification Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for microbial identification Microbial DNA qPCR Assays reveals high sensitivity.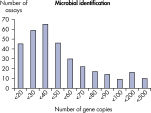 The lower limit of quantification (LLOQ) for virulence factor gene detection Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for virulence factor gene detection Microbial DNA qPCR Assays reveals high sensitivity.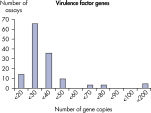 The lower limit of quantification (LLOQ) for antibiotic resistance gene detection Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for antibiotic resistance gene detection Microbial DNA qPCR Assays reveals high sensitivity.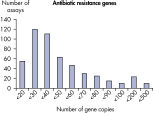 Microbial DNA qPCR Assays are highly specific.
Microbial DNA qPCR Assays are highly specific.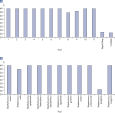 Microbial DNA qPCR Assays display high sensitivity even in complex metagenomic samples.
Microbial DNA qPCR Assays display high sensitivity even in complex metagenomic samples.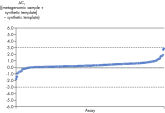
 The lower limit of quantification (LLOQ) for all Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for all Microbial DNA qPCR Assays reveals high sensitivity. The lower limit of quantification (LLOQ) for microbial identification Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for microbial identification Microbial DNA qPCR Assays reveals high sensitivity. The lower limit of quantification (LLOQ) for virulence factor gene detection Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for virulence factor gene detection Microbial DNA qPCR Assays reveals high sensitivity. The lower limit of quantification (LLOQ) for antibiotic resistance gene detection Microbial DNA qPCR Assays reveals high sensitivity.
The lower limit of quantification (LLOQ) for antibiotic resistance gene detection Microbial DNA qPCR Assays reveals high sensitivity. Microbial DNA qPCR Assays are highly specific.
Microbial DNA qPCR Assays are highly specific. Microbial DNA qPCR Assays display high sensitivity even in complex metagenomic samples.
Microbial DNA qPCR Assays display high sensitivity even in complex metagenomic samples.
Principle
Microbial DNA qPCR Assays are designed to detect bacterial 16S rRNA gene and fungal ribosomal rRNA gene sequences for species identification, as well as detecting virulence factor genes and antibiotic resistance genes using PCR amplification primers and hydrolysis-probe detection.
Procedure
The procedure for Microbial DNA qPCR Assays is simple, and can be performed in any laboratory with a real-time PCR instrument. DNA is isolated from the sample using the appropriate QIAamp kit, and then PCR reactions are set up using the appropriate Supplemental Microbial qPCR Mastermix for the PCR instrument. Four separate PCR reactions should be prepared for each sample, including a Positive PCR Control, a No Template Control, and the Microbial DNA Positive Control, as well as the Microbial DNA qPCR Assay. Real-time PCR is performed and data is analyzed using web-based data analysis software or Excel templates.
Applications
Microbial DNA qPCR Assays are highly suited for the detection of bacterial or fungal species, or microbial genes for antibiotic resistance or virulence factors, from a variety of samples including stool, sputum, vaginal swab, sewage, and others.
Supporting data and figures
Limit of detection versus lower limit of quantification.
This chart demonstrates the difference between the limit of detection (LOD) and the lower limit of quantification (LLOQ). The LOD is defined as the lowest concentration at which 95% of the positive samples are detected, whereas the LLOQ is the lowest concentration that falls within the linear range of a standard curve. LOD depends upon the precision of the assay, and requires at least 40 replicates for determination of a positive sample. For the Microbial DNA qPCR Assays, LLOQ is sufficient to determine assay sensitivity.
Resources
Safety Data Sheets (1)
Brochures & Guides (2)
Scientific Posters (1)
Technical Information (2)
Kit Handbooks (1)
Certificates of Analysis (1)
FAQ
What species are detected by the Pan Bacteria 1 and Pan Bacteria 3 Assays?
What sequences are used to design the Microbial DNA qPCR Assays?
What are the storage conditions for the Microbial DNA qPCR products?
What is the difference between LLOQ and LOD?
What is the difference between Positive PCR Control (PPC) and Microbial DNA Positive Control?
Is the Microbial qPCR mastermix used in the Microbial DNA assay and in the Microbial DNA arrays free of genomic bacterial DNA?
Can I multiplex the Microbial DNA qPCR Assays?
What sample types can be tested on the arrays/assays?
Can I measure antibiotic resistance gene expression?
What is the expected amplicon size of the Microbial DNA qPCR Assays?
What is LLOQ?
Can I measure virulence factor gene expression?
Can I use the Microbial DNA-Free Water and Microbial qPCR Mastermix if they have been opened more than 3 times?
How can I calculate the number of bacterial cells that are present in a sample using the Microbial DNA qPCR Assays?
Are the Microbial DNA qPCR Assays wet-lab verified?
Are the assays species-specific?
Which Microbial qPCR Mastermix should I use?
What are the minimum sample requirements for Microbial DNA qPCR kits?
Which probe labels are available for the Microbial DNA qPCR Assays?
What is the sensitivity for the Microbial DNA qPCR kits?

