✓ 연중무휴 하루 24시간 자동 온라인 주문 처리
✓ 풍부한 지식과 전문성을 갖춘 제품 및 기술 지원
✓ 신속하고 안정적인 (재)주문
QIAamp DNA Micro Kit (50)
카탈로그 번호 / ID. 56304
✓ 연중무휴 하루 24시간 자동 온라인 주문 처리
✓ 풍부한 지식과 전문성을 갖춘 제품 및 기술 지원
✓ 신속하고 안정적인 (재)주문
특징
- 고품질 DNA의 신속한 정제
- 유기 추출 또는 알코올 침전물 없음
- 일관되고 높은 수율
- 오염 물질 및 억제제 완전 제거
- QIAcube Connect에서 1~12개 샘플 자동 정제
제품 세부 정보
QIAamp DNA Kit는 조직, 면봉, CSF, 혈액, 체액 또는 소변에서 세척한 세포로부터 실리카 멤브레인을 기반으로 핵산을 정제합니다. 또한, 소량의 신선 또는 냉동 혈액, 조직 및 건조 혈반에서 유전체 DNA와 미토콘드리아 DNA를 정제할 수 있습니다. 조직은 효소로 분해되므로 기계적 균질화가 필요하지 않습니다. 전용 QIAamp DNA Mini QIAcube Kit를 사용하여 QIAcube Connect에서 1~12개 샘플 DNA 정제를 자동화할 수 있습니다. QIAamp DNA Micro Kit 및 QIAamp DNA Mini Kit을 사용하는 DNA 정제도 QIAcube Connect에서 자동화할 수 있습니다.
QIAamp DNA Mini 표준 프로토콜은 또한 TRACKMAN Connected 시스템을 이용해서도 실행할 수 있습니다.
성능
QIAamp DNA Micro Kit는 실리카 기반 막의 선택적 결합 특성과 20~100µL 사이의 유연한 용출 용량이 조합되었습니다. QIAamp DNA Micro Kit을 사용하여 정제한 DNA는 단백질, 핵산분해효소 및 기타 불순물이 없으며 real-time PCR(그림 ' 소량 샘플에서의 효율적인 DNA 정제' 참조) 및 레이저 미세절단(Laser Microdissection, LMD) PCR(그림 ' 레이저 미세절단 PCR' 참조)과 같은 고감도 다운스트림 응용 분야에 사용하기 적합합니다. 정제된 DNA는 단연쇄 반복(Short-Tandem Repeat, STR) 유전형 분석, 단일 염기 다형성(Single-Nucleotide Polymorphism, SNP) 유전형 분석 또는 약리유전체학 연구에도 사용할 수 있습니다.
QIAamp DNA mini Kit는 신속 스핀 컬럼이나 진공 절차를 통해 조직 샘플에서 DNA를 분리하는 과정을 간소화하여 최대 50kb 크기의 DNA를 얻을 수 있습니다(그림 ' 최대 50kb 유전체 DNA 정제' 참조). 이 길이의 DNA는 완전히 변성되며 증폭 효율이 가장 높습니다. 핵산이나 DNA의 수율은 시재료에 따라 달라집니다(표 'QIAamp DNA Mini Kit의 일반적인 수율' 참조). QIAamp DNA Mini Kit을 사용하여 정제된 DNA는 PCR 및 정량 real-time PCR, 서던 블로팅, SNP 및 STR 유전형 분석, 약리유전체학 연구를 포함한 다양한 다운스트림 응용 분야에 사용할 수 있습니다.
| 샘플 | 양 | 총 핵산 수율(µg)* | DNA 수율 (µg)† |
|---|---|---|---|
| 혈액 | 200µL | 4~12 | 4~12 |
| 백혈구 연층 | 200µL | 25~50 | 25~50 |
| 세포 | 106 | 20~30 | 15~20 |
| 간 | 25mg | 60~115 | 10~30 |
| 뇌 | 25mg | 35~60 | 15~30 |
| 폐 | 25mg | 25~45 | 5~10 |
| 심장 | 25mg | 15~40 | 5~10 |
| 신장 | 25mg | 40~85 | 15~30 |
| 비장 | 10mg | 25~45 | 5~30 |
QIAamp DNA Mini QIAcube Kit는 최대 50kb 크기의 DNA를 생산합니다. 이 길이의 DNA는 완전히 변성되며 증폭 효율이 가장 높습니다. 정제된 DNA는 다음을 포함한 다양한 응용 분야에서 사용될 수 있습니다.
- 바이러스 연구
- 세균 연구
- 균류 연구
- 암 연구
- 인간 유전체 시험 연구
- 친자 확인 검사
- 법의학 분석
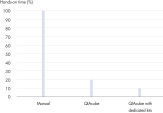
QIAamp DNA Mini QIAcube Kit는 신속 스핀 컬럼 절차를 통해 조직 샘플에서 DNA를 분리하는 과정을 간소화합니다. 이 키트에는 QIAamp 스핀 컬럼과 용출 튜브가 사전에 장착되어 있는 로터 어댑터가 포함되어 있어 편의성이 더욱 높으며 시간도 절약됩니다(그림 ' 상당한 시간 절약' 참조).
원리
QIAamp DNA 정제 절차 중 오염 물질이 통과하는 동안 QIAamp MinElute 또는 QIAamp 실리카겔 멤브레인에 DNA가 특이적으로 결합합니다. 페놀-클로로포름 추출은 필요하지 않습니다. 2가 양이온 및 단백질과 같은 PCR 억제제는 효율적인 2단계 세척을 통해 완전히 제거되며, 키트와 함께 제공되는 정제수 또는 완충액에서 순수한 DNA가 용출됩니다.
QIAamp DNA 기술은 인체 조직 샘플에서 PCR 및 블로팅 절차에 바로 사용할 수 있는 유전체, 미토콘드리아, 박테리아 또는 바이러스 DNA를 생산합니다.
QIAamp 샘플 준비 기술은 라이선스를 완벽히 취득하여 QIAamp에서 정제된 핵산을 어떤 분자 분석이나 기타 다운스트림 응용 분야에도 특허 침해 위험 없이 사용할 수 있습니다.
절차
QIAamp DNA Mini Kit에 있는 최적화된 완충액과 효소가 샘플을 용해하고, 핵산을 안정화하며, QIAamp 멤브레인에 대한 선택적 DNA의 흡착을 증강합니다. 알코올이 첨가되고 용해물이 QIAamp 스핀 컬럼에 로드됩니다.
세척 완충액을 사용하여 불순물을 제거하고 바로 사용할 수 있는 순수한 DNA가 물 또는 저염 완충액에 용출됩니다.
조직은 효소로 분해되므로 기계적 균질화가 필요하지 않습니다. 편리한 스핀 컬럼 방식은 직접 준비하는 데 20분밖에 걸리지 않습니다(용해 시간은 샘플 공급원에 따라 다름).
샘플은 마이크로 원심분리기를 사용해서 처리하거나, 아니면 혈액이나 기타 체액을 처리하는 경우에는 QIAvac 24 Plus를 사용할 수 있습니다. 또한, QIAamp DNA Mini Kit는 엄격한 용해 절차를 채택하여 박테리아나 기생충에서 유전체 DNA를 정제하는 데 이상적입니다.
수작업 시간을 더욱 단축하기 위해, QIAcube에서의 유전체 DNA 정제를 자동화할 수 있습니다. QIAamp DNA 액세서리 세트 A는 QIAcube와 12 x QIAamp DNA Mini Kits (50)를 사용하여 유전체, 미토콘드리아, 박테리아, 기생충 또는 바이러스 DNA를 분리하기 위한 모든 완충액 및 시약을 제공합니다.
진공 공정
DNA 정제를 더욱 빠르고 편리하게 하기 위해 원심분리 대신 진공 방식을 사용해 혈액이나 기타 체액을 처리할 수 있습니다. QIAamp mini 스핀 컬럼은 VacValves와 VacConnectors를 사용하는 QIAvac 24 Plus 매니폴드에 들어갑니다.
샘플 유량이 크게 다를 경우에 일관된 진공을 보장하려면 VacValve를 사용해야 합니다. 교차 오염을 방지하기 위해 일회용 VacConnector를 사용합니다. VacConnectors를 사용하면 QIAvac Luer 어댑터를 사용하는 QIAvac 6S에서도 QIAamp 스핀 절차를 수행할 수 있습니다.
QIAcube Connect에서 자동 DNA 분리
전용 QIAamp DNA Mini QIAcube Kit는 QIAcube 기기용 로터 어댑터에 미리 장착된 스핀 컬럼과 용출 튜브를 사용하여 설계되었으며, 로터 어댑터를 잘못 장착하여 발생할 수 있는 오류 위험을 제거합니다. 전용 키트는 QIAcube 요구 사항에 맞게 제작되어 낭비를 줄입니다.
QIAcube Connect의 간단한 프로토콜(용해, 결합, 세척 및 용출)과 자동화 프로토콜은 최소한의 사용자 상호 작용만 필요합니다.
수상 경력이 있는 QIAcube Connect는 첨단 기술을 사용하여 QIAGEN 스핀 칼럼을 자동으로 처리하며, 자동화된 저처리량 시료 전처리 과정을 실험실 워크플로우에 원활하게 통합할 수 있도록 지원합니다. 정제 절차의 모든 단계는 전면 자동화되어 있으며, 한 번 실행에 최대 12개의 샘플을 처리할 수 있습니다. QIAcube Connect와 전용 QIAamp DNA Mini QIAcube 키트로 DNA를 빠르고 쉽고 편리하게 정제할 수 있습니다.
응용 분야
QIAamp DNA Micro Kit 절차는 소량의 혈액, 혈액 카드, 소변, 레이저 미세절제를 포함한 소량 조직 샘플을 포함하는 광범위한 샘플 재료에 적합합니다.
QIAamp DNA Mini Kit는 근육, 간, 심장, 뇌, 골수 및 기타 조직, 면봉(구강, 눈, 비강, 인두 및 기타), 뇌척수액, 혈액, 체액 및 소변에서 세척한 세포를 포함한 가장 일반적으로 사용되는 인체 조직 샘플에서 DNA를 정제하는 데 이상적입니다. 최대 25mg의 조직 또는 최대 200µL의 체액에서 20분 이내에 DNA를 정제하고, 50~200µL에서 용출할 수 있습니다.
QIAamp DNA Mini QIAcube Kit는 QIAcube Connect에서 유전체, 미토콘드리아, 기생충 또는 바이러스 DNA를 자동으로 분리하는 데 사용됩니다. 샘플 출처로는 조직, 구강 면봉, CSF, 소변에서 세척한 세포가 포함됩니다.
| 특징 | QIAamp DNA Micro Kit | QIAamp DNA Mini Kit |
|---|---|---|
| 응용 분야 | Real-time PCR, STR 분석, LMD-PCR | PCR, 서던 블로팅 |
| 용출량 | 20~100µL | 50~200µL |
| 형식 | 스핀 컬럼 | 스핀 컬럼 |
| 주 샘플 유형 | 전혈 | 전혈, 조직, 세포 |
| 처리 | 수동(원심분리 또는 진공) | 수동(원심분리 또는 진공) |
| total RNA, miRNA, poly A+ mRNA, DNA 또는 단백질 정제 | 유전체 DNA, 미토콘드리아 DNA | 유전체 DNA, 미토콘드리아 DNA, 박테리아 DNA, 기생충 DNA, 바이러스 DNA |
| 샘플양 | 1~100µL | 200µL/25mg/5 x 106 |
| 기술 | 실리카 기술 | 실리카 기술 |
| 실행당 또는 준비당 시간 | 30분 | 20분 |
| 수율 | <3µg | 4~30µg |
지원되는 데이터 및 수치
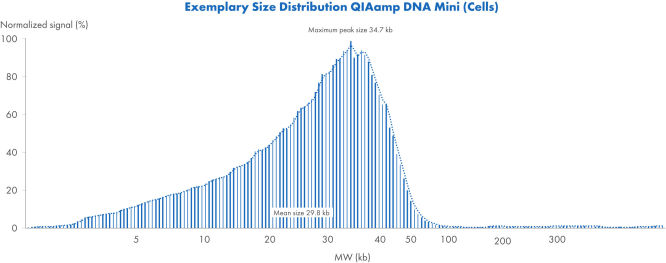
QIAamp DNA Mini Kit(세포)를 사용한 크기 분포 예시
QIAamp DNA Mini Kit를 사용하여 세포에서 분리한 DNA 크기 분포 예시. 샘플은 Femto Pulse System을 사용하여 분석하고 신호는 최대 피크로 정규화했습니다.