RNeasy Fibrous Tissue Mini Kit
从富含纤维的组织中纯化至多100 µg的总RNA
从富含纤维的组织中纯化至多100 µg的总RNA
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 74704
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
RNeasy Fibrous Tissue Kits适用于从纤维丰富组织中提取RNA,RNeasy Fibrous Tissue操作流程能简单有效的提取高品质总RNA(参见" High-quality RNA from fiber-rich tissues"),纯化的RNA适用于多种下游应用,包括芯片分析和real-time RT-PCR (参见 " Real-time gene expression analysis in a variety of fibrous tissues" 和" LightCycler analysis of high-quality RNA from heart")。与其他硅胶方法相比,RNeasy Fibrous Tissue Mini Kit从纤维丰富组织中提取的RNA产量更高(参见下表)。
| 大鼠组织(10 mg) | 使用RNeasy Fibrous Tissue Mini Kit 得到的RNA产量* (µg) | 使用硅胶试剂盒得到的RNA产量*(µg)(供应商AV) | 使用硅胶试剂盒得到的RNA产量*(µg)(供应商R) | 使用硅胶试剂盒得到的RNA产量*(µg)(供应商S) |
|---|---|---|---|---|
| 心脏 | 8.2 | 3.5 | <0.5 | 2.5 |
| 肌肉 | 5.3 | 0.5 | 2.0† | 2.2 |
| 皮肤 | 6.0 | 未确定 | 未确定 | 未确定 |
| 食道 | 13.9 | 未确定 | 未确定 | 未确定 |
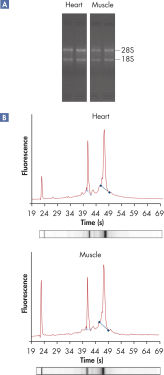
RNeasy技术具有异硫氰酸胍裂解的严谨性以及硅胶膜纯化的速度和纯度,简化了总RNA的纯化流程。RNeasy Fibrous Tissue Kits适用于处理纤维丰富的组织,例如骨骼肌、心脏和主动脉。因为含有大量的收缩性蛋白、连接组织和胶原蛋白,这些组织非常难裂解。RNeasy Fibrous Tissue操作流程将蛋白酶K和RNase-free DNase消化步骤结合起来,在RNA分离的同时能降解蛋白和基因组DNA,纯化得到的高品质总RNA的产量更高。RNeasy技术适用于大于200 nt的RNA,因此RNA产物不包括5S rRNA、tRNA或其他小分子量RNAs,产物占细胞内总RNA的15–20%。
0.5–30 mg纤维组织样本在异硫氰酸胍缓冲液中裂解。裂解液被稀释后,样本经蛋白酶K处理。离心使碎片沉淀。然后加入乙醇使裂解液澄清,之后RNA结合到RNeasy硅胶膜上。在RNeasy离心柱中加入DNase去除残留的DNA。DNase和其他污染物被洗涤后,使用150–500 µl不含RNase的水即可洗脱得到至多100 µg的高品质总RNA(参见" RNeasy Fibrous Tissue Midi procedure")。
RNeasy Fibrous Tissue Kits从纤维丰富的样本中纯化得到的高品质总RNA,适用于多种下游应用,包括芯片分析和real-time RT-PCR。
| Features | Specifications |
|---|---|
| Applications | PCR, real-time PCR, microarray |
| Elution volume | 30–100 µl |
| Format | Spin column |
| Time per run or per prep | 55 minutes |
| Purification of total RNA, miRNA, poly A+ mRNA, DNA or protein | RNA |
| Processing | Manual |
| Sample amount | 0.5–30 mg |
| Technology | Silica technology |
| Main sample type | Fibre-rich tissue samples |
| Yield | 5–12 µg |