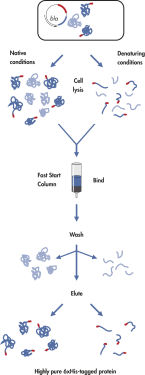
To optimize the expression of a given recombinant protein, a time-course analysis of the level of protein expression in the induced culture is recommended. Intracellular protein content is often a balance between the amount of soluble protein in the cells, the formation of inclusion bodies, and protein degradation. By checking the 6xHis-tagged protein present at various times after induction in the soluble and insoluble fractions, the optimal induction period can be established, and the bacterial culture can be harvested at this time. It may be useful to perform plasmid Mini preparations on culture samples during the time-course to enable monitoring of plasmid (expression construct) maintenance.
Below, you can see an example of a time course of recombinant protein expression using the QIAexpress System. You can find this information also in the Section 'Expression in E. coli' in the QIAexpressionist Handbook. The handbook is an important resource for useful background information and protocols. For instructions on how to isolate protein from the soluble and insoluble fractions of induced cultures please see Protocol 14. "Protein minipreps of 6x His-tagged proteins from E. coli under native conditions" and Protocol 19. "6xHis-tagged protein minipreps under denaturing conditions."

Time course of expression using the QIAexpress System. Expression of 6xHis-tagged DHFR was induced with 1 mM IPTG. Aliquots were removed at the times indicated and purified on Ni-NTA Agarose under denaturing conditions. Proteins were visualized by Coomassie staining. Yields per liter culture were 2.8, 5.5,12.3, 33.8, and 53.9 mg, respectively. ■A Crude cell lysate; ■B purification with Ni-NTA. 1: flow-through, 2 & 3: first and second eluates; M: markers; C: noninduced control.