RT2 PreAMP Pathway Primer Mixes
用于RT² Profiler PCR Arrays分析之前扩增cDNA模板
用于RT² Profiler PCR Arrays分析之前扩增cDNA模板
Cat. No. / ID: 330241
RT² PreAMP cDNA Synthesis Kit和RT² PreAMP Pathway Primer Mixes是创新技术,能够用低至1 ng的总RNA进行基因表达分析。专有技术的扩增过程增加了用于PCR芯片分析的cDNA的量。起始样本包括穿刺活检、激光捕获显微解剖样本、干细胞团或胚胎体及流式细胞分选仪(FACS)制备的细胞。专门设计的引物混合液适用于人类、小鼠或大鼠的Custom RT² Profiler PCR Arrays。
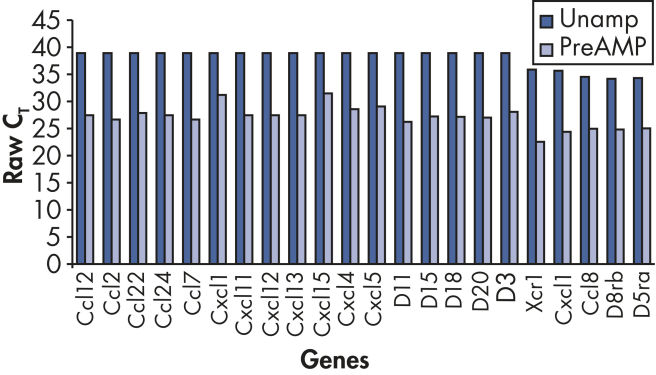
使用RT² PreAMP Pathway Primer Mix进行预扩增之后,可检测的基因更多(参见" Increased positive call rate")。
扩增过程无偏向性,回归分析评估发现,预扩增的与未扩增的cDNA之间能产生高度可比较的ΔCT值(参见" Unbiased amplification process")。
在预扩增与未扩增样本之间,基因表达倍数变化有高度相关性(参见" Faithfully amplified biology")。
首先使用RT² PreAMP cDNA Synthesis Kit同时反转录最多12 种不同的RNA样本为cDNA。然后将cDNA进行预扩增用于特定的基因列表。每一个反转录反应用4种RT² PreAMP Pathway Primer Mixes进行扩增,能够在4个特定PCR 芯片上进行基因表达分析。RT² PreAMP cDNA Synthesis Kit中含有Side Reaction Reducer,可去除预扩增所产生的引物残留,能在RT² Profiler PCR Arrays进行准确检测。把预扩增的模板与仪器特异性、即用型RT² SYBR® Green Mastermix混合, 便完成PCR芯片过程。