Cat. No. / ID: 34362
Features
- Expressed His-tag optimized for removal by TAGZyme enzyme
- Efficient His-tag removal: >95% in just 30 minutes at 37°C
- High-level expression of N-terminally His-tagged proteins
- High-purity end products
- Complete removal of contaminants by Ni-NTA method
Product Details
The TAGZyme Enzyme DAPase includes sufficient enzyme for highly specific and efficient His-tag removal from up to 10 mg His-tagged protein. The TAGZyme system can be used for His-tag removal from proteins containing an intrinsic DAPase stop point expressed using the TAGZyme pQE-2 vector.
Performance
TAGZyme DAPase Enzyme efficiently removes dipeptides sequentially from N-terminal His tags up to the "stop point" expressed using TAGZyme pQE-2 vector.
Principle
His-tagged recombinant proteins have become valuable tools in studying protein structure and function. The small size and low immunogenicity of the His tag means that its removal is not usually required. However, a protein product free from vector-derived amino acids is preferred for some applications, such as structure-determination studies by X-ray or NMR, or the production of therapeutics.
The TAGZyme pQE-2 vector is suitable for proteins containing an intrinsic DAPase stop point.
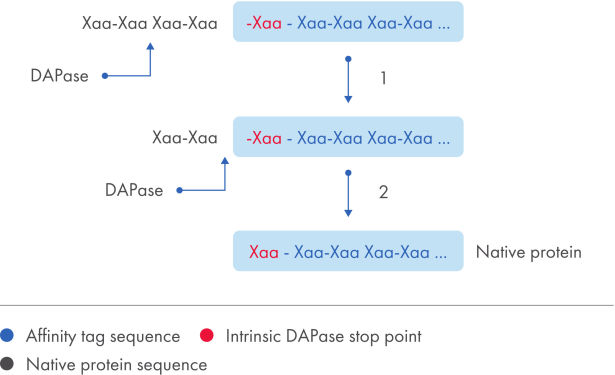
The TAGZyme system removes N-terminal His tags from recombinant proteins with high specificity and efficiency. DAPase enzyme is used to sequentially cleave off dipeptides from the N-terminus of the purified, His-tagged protein (see figure "His-tag removal”). Digestion is halted when the enzyme reaches a “stop point”, an amino acid motif that cannot serve as a substrate (see table "DAPase stop points").
DAPase stop points
| Amino acid | DAPase stop point (↓) sequence* |
|---|---|
| Lysine (Lys, K) | Xaa-Xaa...Xaa-Xaa ↓ Lys-Xaa... |
| Arginine (Arg, R) | Xaa-Xaa...Xaa-Xaa ↓ Arg-Xaa... |
| Proline (Pro, P) | Xaa-Xaa...Xaa-Xaa ↓ Xaa-Xaa-Pro-Xaa... |
| Proline (Pro, P) | Xaa-Xaa...Xaa-Xaa ↓ Xaa-Pro-Xaa-Xaa... |
| Glutamine (Gln, Q)† | Xaa-Xaa...Xaa-Xaa ↓ Gln-Xaa... |
| Isoleucine (Ile, I) | Xaa-Xaa...Xaa-Xaa ↓ Xaa-Ile-Xaa-Xaa... |
See figures
Procedure
With recombinant proteins that contain intrinsic stop points, expression using the TAGZyme pQE-2 vector allows complete and efficient removal of the N-terminal His tag irrespective of the cloning site of the DNA insert (see figure "His-tag removal"). After incubation with DAPase enzyme, the reaction mixture is subjected to subtractive immobilized-metal affinity chromatography (IMAC) using a Ni-NTA matrix (see figure "Purification of detagged proteins"). His-tag fragments and TAGZyme DAPase Enzyme (which carries a C-terminal 6xHis tag) bind to the matrix, and pure, detagged target protein is recovered in the flow-through fraction.
See figures
Applications
The TAGZyme system offers specific cleavage, the use of recombinant reagents and the complete removal of all contaminants, making it the method of choice for the production of His-tag-free proteins for applications including:
- Protein structure determination by NMR or X-ray crystallography
- Production of therapeutic proteins
Supporting data and figures
His-tag removal.
Schematic summary of the overall cleavage strategy using TAGZyme enzymes. DAPase enzyme cleavage of a N-terminal His tag from a protein containing a natural stop point to obtain the mature target protein.