✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
dPCR Microbial DNA Detection Assays
Cat no. / ID. 250207
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Useful links
Below you can find a set of references
Features
- Predesigned assays for the detection of microbial species, virulence genes or antibiotic resistance genes
- Assays for more than 800 targets
- Custom assay design tool for bacterial, fungal or viral targets available on GeneGlobe
- Dye selection enables multiplexing of up to 12 targets per reaction
- Simple and fast dPCR workflow on the QIAcuity
- Combine microbial DNA and viral RNA targets in one reaction using the QIAcuity OneStep Advanced Probe Kit
Product Details
Microbial identification and profiling are of interest to many fields including human health, food and feed testing and environmental testing. Microbial identification determines the presence or absence of a microbe in a sample, while microbial profiling determines its relative expression under two or more experimental conditions, thus requiring a reference sample and a normalizer.
Our dPCR Microbial DNA Detection Assays target bacterial, fungal, parasitic, viral, antibiotic resistance or virulence factor genes. For bacteria, the assays target the 16S rRNA gene, and for fungi, they target ribosomal RNA genes. The portfolio consists of over 700 different assays, 200 of which are dPCR wet-lab tested. For targets not covered by our portfolio of predesigned microbial dPCR assays, we offer an easy to use custom assay design tool for bacterial, fungal and viral targets.
Each assay consists of a primer pair and a hydrolysis probe with a configurable fluorophore dye. The available fluorophores include FAM, HEX, ROX, TAMRA, Cy5 and ATTO700, and these can be mixed and matched to support analysis of up to twelve different targets in one multiplex dPCR reaction. We also offer wet-lab tested dPCR 5-plex assay bundles for analyzing common targets of interest; see Applications below for more details. For the dPCR reaction, the assay is combined with the QIAcuity Probe PCR Kit (DNA targets) or the QIAcuity OneStep Advanced Probe Kit (RNA targets) or the QIAcuity High Multiplex Probe PCR Kit (for more than five targets in one reaction).
The assay works in conjunction with the QIAcuity Digital PCR System and the QIAcuity Nanoplates.
Would you like to learn more about this product and be contacted by one of our dPCR specialists? Sign in here, and we will get in touch with you shortly.
Performance
Accurate and precise detection
The dPCR Microbial DNA Detection Assays and the QIAcuity dPCR System enable accurate digital PCR-based quantification. Analysis of NIST reference material showed the expected concentration of input template. See figure Quantification of the NIST reference standard 8376 using the gDNA from Shigella sonnei for more details.
Performance in multiplex
Multiplex dPCR-based detection of microbial targets offers a flexible setup of small panels in fewer reactions, thus conserving precious samples. Multiplexing also boosts the analysis throughput, as it requires fewer Nanoplate wells and increases the number of samples that can be analyzed in each run. Our dPCR Microbial DNA Detection Assays can be combined for multiplex analysis.
You can combine your own assays for multiplexing by ordering single assays with different dyes (FAM, HEX, TAMRA, ROX, Cy5 and ATTO700). Or, you can choose from our offering of 5-plex assay bundles, which have already wet-lab tested in dPCR by our scientists. Other combinations of the >700 predesigned assays should be verified in your own lab.
A comparison between singleplex and multiplex analysis using the dPCR Microbial DNA Detection Assays shows similar and highly precise quantification for all targets. In addition to precise target quantification, a 12-plex dPCR run also shows highly specific detection of different microbial water pathogens.
Principle
The predesigned Microbial DNA Detection Assays and the Custom dPCR Microbial Assays are intended for use in nanoplate digital PCR. Each assay is based on an endpoint PCR amplification of a species-specific genetic region of the relevant microbe or a region of an individual microbial gene. The amplified product is detected using target-specific fluorescent hydrolysis probes, which improves the specificity of the assay.
The dPCR Microbial DNA Detection Assays for detecting bacterial species target the ribosomal RNA genes – mainly the 16S ribosomal RNA gene – and were designed using the GreenGenes database for 16S sequences and type strain DNA sequences deposited at NCBI. Assays for fungal, viral and metazoan species target different target-specific genetic regions, including ribosomal RNA genes and other individual marker genes, each deposited at NCBI. Various databases were used for the design of assays for antibiotic resistance genes (e.g., lahey.org, ARDB, etc.) and virulence factor genes (e.g., VFDB).
The Custom dPCR Microbial Assays are created with our custom assay design software which is based on a sophisticated and thoroughly tested algorithm developed specifically for microbial targets. It ensures each assay design meets stringent performance criteria to deliver robust, high-quality assays with optimal sensitivity and specificity.
The principle of the dPCR reaction in the nanoplates is described here.
Procedure
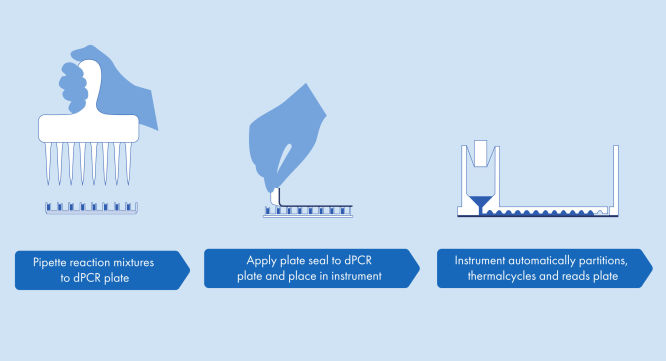
The dPCR Microbial DNA Detection Assay and the Custom dPCR Microbial Assays protocol is simple and can be performed in any laboratory with the QIAcuity dPCR instrument. DNA is isolated from the sample and is added to the ready-to-use QIAcuity Probe Mastermix and Microbial DNA-Free Water (UCP water). This mix is aliquoted into each well of the dPCR pre-plate, which contains pre-dispensed sets of primers and hydrolysis probes. The reaction mixes are transferred from the pre-plate to the wells of a dPCR Nanoplate, which is then sealed and transferred to the QIAcuity dPCR instrument (see figure A simple and rapid plate-based workflow). The partitioning, cycling and imaging steps are fully automated by the QIAcuity dPCR instrument following the preset parameters. Depending on the cycling protocol, the results of the dPCR run can be analyzed in the QIAcuity Software Suite after ~2 hours (see figure Microbial dPCR in about 2 hours with minimal hands-on time).
Applications
The dPCR Microbial DNA Detection Assays are highly suited for the detection of bacterial, fungal and viral species and microbial antibiotic resistance or virulence factor genes. A variety of samples can be analyzed, including stool, sputum, vaginal swab, sewage and others.
Additionally, the Custom dPCR Microbial assays design tool allows to seamlessly design assays for any microbial (bacteria, fungi, viruses) target of interest not covered by our predesigned dPCR Microbial DNA Detection Assays.
dPCR wet-lab tested 5-plex bundles
| Bundle ID | Application Field | Assay Catalog Numbers and Fluorophore* |
Targets (NCBI Taxonomy ID) |
| WW-001 | Wastewater 1 | DMA00278-F DMA00291-R DMA00340-H DMA00192-T DMA00344-C |
Pseudomonas aeruginosa (287) Salmonella enterica (28901) Vibrio cholerae (666) Legionella pneumophila (446) Yersinia enterocolitica (630) |
| WW-002 | Wastewater 2 | DMA00340-H DMA00344-R DMA00199-C DMA00192-T DMA00710-F |
Vibrio cholerae (666) Yersinia enterocolitica (630) Listeria monocytogenes (1639) Legionella pneumophila (446) Human corona virus SARS-CoV-2 (2697049) |
| WW-003 | Wastewater 3 | DMA00109-F DMA00192-H DMA00340-T DMA00194-R DMA00344-C |
Clostridium perfringens (1502) Legionella pneumophila (446) Vibrio cholerae (666) Leptospira alexanderi (100053) Yersinia enterocolitica (630) |
| HM-001 | Human Microbiome 1 | DMA00148-F DMA00143-T DMA00024-R DMA00003-C DMA00150-H |
Faecalibacterium prausnitzii (853) Eubacterium rectale (39491) Akkermansia muciniphila (239935) Acidaminococcus fermentans (951) Finegoldia magna (1260) |
| PB-001 | Probiotics 1 | DMA00177-F DMA00185-T DMA00061-C DMA00137-R DMA00320-H |
Lactobacillus acidophilus (1579) Lactiplantibacillus plantarum (1590) Bifidobacterium bifidum (1681) Enterococcus faecium (1352) Streptococcus salivarius (1304) |
| RG-001 | Resistance Genes 1 | DMA00566-F DMA00542-H DMA00576-T DMA00574-R DMA00575-C |
Fluoroquinolone resistance gene QnrS Class D beta-lactamase OXA-10 group Vancomycin resistance gene vanB Tetracycline efflux pump gene tetA Tetracycline efflux pump gene tetB |
| RG-002 | Resistance Genes 2 | DMA00557-T DMA00528-R DMA00548-F DMA00587-H DMA00553-C |
Fluoroquinolone resistance geneQepA Class B beta-lactamase blaVIM-1 group Class D beta-lactamase OXA-48 group Sulfonamide resistance gene sul1 (43904) Class D beta-lactamase OXA-58 group |
| VG-001 | Virulence Genes 1 | DMA00614-F DMA00635-T DMA00677-H DMA00680-R DMA00597-C |
Minor fimbrial subunit (fimH) Gamma-hemolysin component B (hlgB) Shiga-like toxin 1 subunit B encoded within prophage CP-933V Shiga toxin subunit B; receptor binding subunit Accessory cholera enterotoxin (ace) |
| CP-001 | Cannabis Production 1 | DMA00278-F DMA00302-H DMA00365-R DMA00199-C |
Pseudomonas aeruginosa (287) Staphylococcus aureus (1280) Aspergillus niger (5061) Listeria monocytogenes (1639) |
| HM-002 | Human Microbiome 2 | DMA00271-F DMA00017-T DMA00049-C DMA00142-H DMA00317-R |
Prevotella oralis (28134) Actinomyces viscosus (1656) Bacteroides fragilis (817) Eubacterium infirmum (56774) Streptococcus oralis (1303) |
| RG-003 | Resistance Genes 3 | DMA00579-F DMA00577-T DMA00573-R DMA00502-H DMA00559-C |
aac(6')-Ib vanC oprm QnrB-1 group CTX-M-1 Group |
| VG-002 | Virulence Genes 2 | DMA00664-F DMA00677-H DMA00678-T DMA00642-R DMA00680-C |
ply stx1B stx2A invA stxB |
| VG-003 | Virulence Genes 3 | DMA00688-F DMA00668-H DMA00596-T DMA00597-R DMA00611-C |
wbkA ptxA ace (E. faecalis) ace (V. cholerae) efaA |
* F: FAM, H: HEX, T: TAMRA, R: ROX or C: Cy5
Supporting data and figures
A simple and rapid plate-based workflow
Partitioning, thermocycling and imaging are all integrated into a single fully automated instrument, delivering results in under two hours. Nanoplate formats are amenable to front-end automation for sample preparation, further reducing hands-on time.