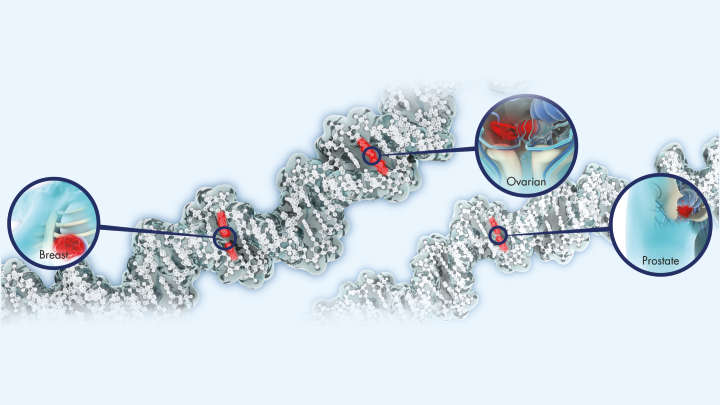
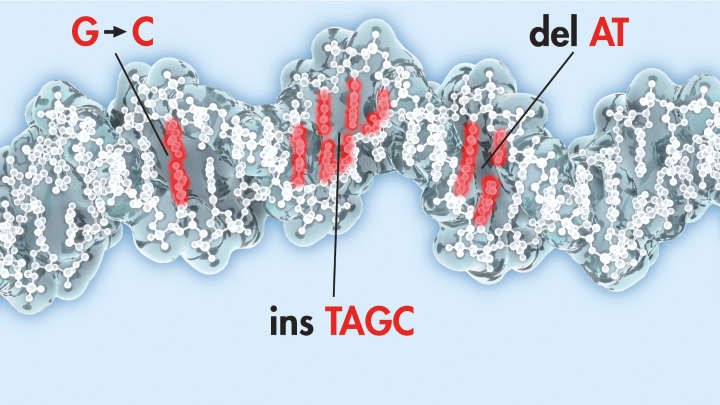
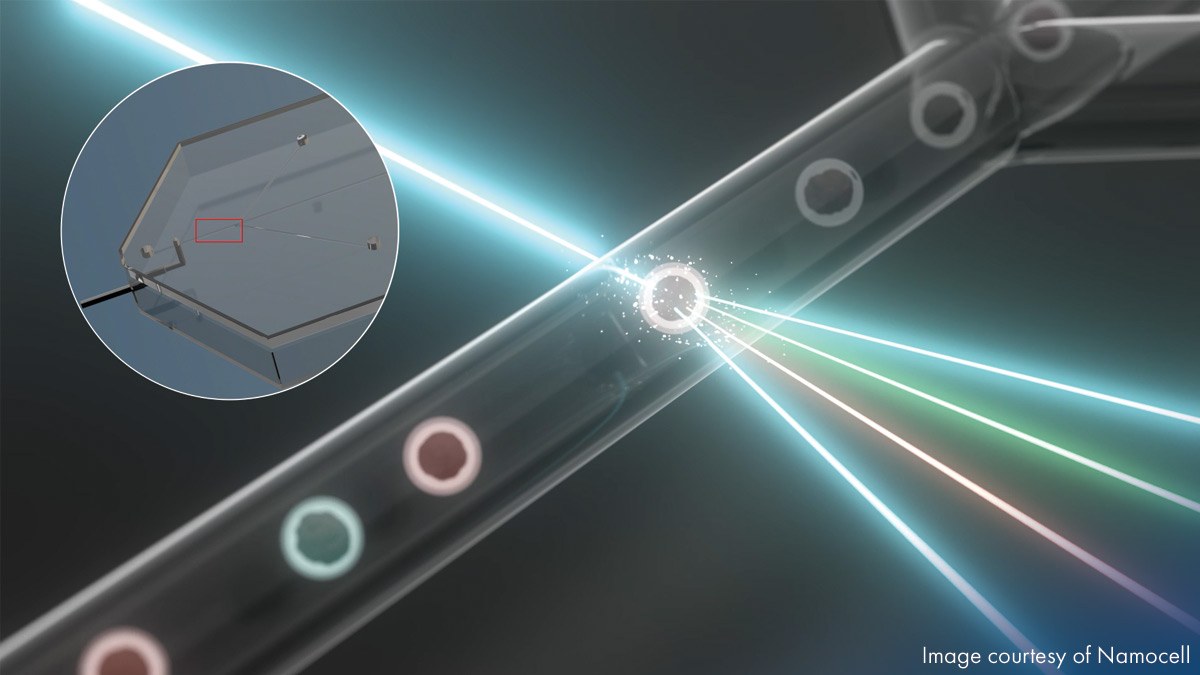
DNA sequencing
Amplicon sequencing
Outstanding sequencing metrics
Uniformity: Standard panels typically achieve uniformity of 99.5% at 0.2x of mean coverage and 96% at 0.5x of mean coverage.
Sensitivity: Gain higher confidence in calling low-frequency DNA variants. Standard panels can obtain over 90% sensitivity for 1% NA12878 SNP and indel on A typical coding region with false positives of less than 15 per megabase region.
More multiplexing: Products from other suppliers offer 96 sample multiplexing. QIAseq does not stop there: 384 samples can be multiplexed using indices specific to QIAseq.
Featured success story
Genomic insights powered by QIAseq
QIAseq solutions are based on the following innovations to minimize bias and maximize genome coverage, uniformity and precision.
Reduce sample mix-up and maximize throughput
Whole exome sequencing
Sample to disease insights in record time
One optimized exome sequencing pipeline with reduced turnaround times to get you from sample to standardized variant classification and interpretation.