QIAexpress Type IV Kit
N-말단 His-tagged 단백질의 고수준 발현 및 원스텝 정제에 사용
N-말단 His-tagged 단백질의 고수준 발현 및 원스텝 정제에 사용
✓ 연중무휴 하루 24시간 자동 온라인 주문 처리
✓ 풍부한 지식과 전문성을 갖춘 제품 및 기술 지원
✓ 신속하고 안정적인 (재)주문
Cat. No. / ID: 32149
✓ 연중무휴 하루 24시간 자동 온라인 주문 처리
✓ 풍부한 지식과 전문성을 갖춘 제품 및 기술 지원
✓ 신속하고 안정적인 (재)주문
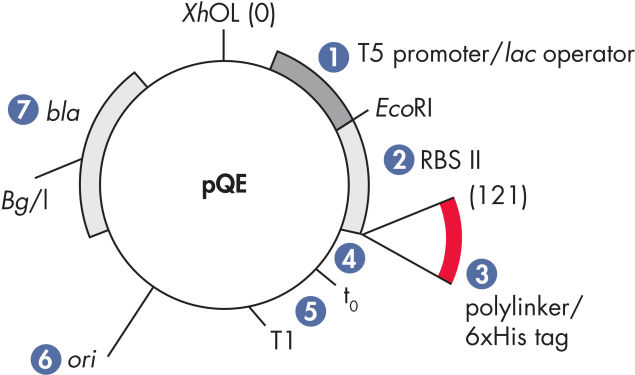
QIAexpress pQE 벡터는 강력한 파지 T5 promoter(E. coli RNA 중합효소에 의해 인식됨)와 이중 lac operator 억제 모듈을 결합하여 E. coli에서 엄격하게 조절된 고수준의 재조합 단백질 발현을 제공합니다. 고수준의 lacrepressor가 존재하면 단백질 합성이 효과적으로 차단되고 세포 독성 구조체의 안정성이 향상됩니다. pQE 벡터(그림 pQE Vector 및 표 참조)를 사용하면 재조합 단백질의 N-말단 또는 C-말단에 His tag를 넣을 수 있습니다.
| 구성 요소 | 설명 |
|---|---|
| 1. 최적화된 promoter/operator 구성 요소 | 파지 T5 promoter와 두 개의 lac operator 염기서열로 구성되며, 이는 lac repressor의 결합 가능성을 높이고 강력한 T5 promoter의 효율적인 억제를 보장합니다 |
| 2. 합성 리보솜 결합 부위 RBSII | 효율적인 번역을 위해 사용 |
| 3. His-tag 코딩 염기서열 | 5' 또는 3'에서 polylinker 클로닝 영역까지 |
| 4. 번역 중지 코돈 | 모든 판독 프레임에서 발현 구조체를 편리하게 준비할 수 있습니다 |
| 5. 두 가지 강력한 전사 종결 인자 | 파지 람다의 t0, E. coli rrnB 오페론의 T1을 사용하여 연속 판독 전사를 방지하고 발현 구조의 안정성을 보장합니다 |
| 6. ColE1 복제 기점 | pBR322 유래 |
| 7. 베타 락타마제 유전자(bla) | 암피실린 내성 부여 |
관심 단백질을 코딩하는 삽입물을 적절한 구조체로 복제하고 발현에 적합한 E. coli 균주로 변형합니다. IPTG를 추가하면 발현이 유도됩니다. QIAexpress Type IV Kit는 E. coli로 형질 전환하거나, 곤충 세포에서 재조합 단백질 발현을 위한 셔틀 벡터로 사용하거나, 포유류 세포에 감염시킬 수 있습니다.
QIAexpress Expression System은 다음을 포함한 많은 애플리케이션에 적합한 고수준 단백질 발현을 제공합니다.
| Features | Specifications |
|---|---|
| Applications | 단백질체학 |
| Yield | 총단백질의 <50% |
| Special feature | 원스텝 정제를 위한 다용도, 완벽한 시스템 |
| Tag | 6xHis tag |
| Processing | 수동 |
| Start material | 세포 용해물 |
| N- or C-terminal tag | N-말단 태그 |