Ni-NTA Agarose
중력류 크로마토그래피에 의한 His 태그 단백질 정제에 사용
중력류 크로마토그래피에 의한 His 태그 단백질 정제에 사용
✓ 연중무휴 하루 24시간 자동 온라인 주문 처리
✓ 풍부한 지식과 전문성을 갖춘 제품 및 기술 지원
✓ 신속하고 안정적인 (재)주문
Cat. No. / ID: 30210
✓ 연중무휴 하루 24시간 자동 온라인 주문 처리
✓ 풍부한 지식과 전문성을 갖춘 제품 및 기술 지원
✓ 신속하고 안정적인 (재)주문
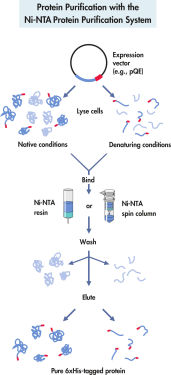
Ni-NTA Agarose는 His 태그가 있는 재조합 단백질을 정제하는 데 사용하는 친화성 크로마토그래피 기질입니다. His 태그의 히스티딘 잔류물은 고정화 니켈 이온의 배위 영역의 빈 위치에 높은 특이성과 친화성으로 결합됩니다. 투명 세포 용해물은 기질로 들어갑니다. His 태그 단백질은 결합되고 다른 단백질은 기질을 통과합니다. His 태그 단백질은 세척 후 기본 또는 변성 조건에서 완충액에 용출됩니다.
Ni-NTA Agarose는 Sepharose CL-6B 받침대에 결합된 Ni-NTA를 제공하며 높은 결합력과 최소한의 비특이적 결합을 제공합니다(그림 기본 조건에서의 단일 단계 정제 참조). 이 물질은 대부분의 배치 및 컬럼 정제 규모에서 다루기 좋은 성질을 가지고 있습니다. Ni-NTA Agarose는 별도로 사용하거나 E. coli에 있는 His 태그 단백질의 효율적인 발현 및 정제를 위한 종합 키트인 QIAexpress Kit의 구성 요소로 사용할 수 있습니다.
QIAexpress Ni-NTA Protein Purification System은 6개 이상의 히스티딘 잔류물(연속 또는 교대)의 친화성 태그(His 태그)를 포함하는 단백질에 대한 특허받은 Ni-NTA(nickel-nitrilotriacetic acid) 수지의 뛰어난 선택성을 기반으로 합니다. 이 기술을 사용하면 기본 또는 변성 조건의 모든 발현 시스템에서 거의 모든 His 태그 단백질을 한 번에 정제할 수 있습니다. 니켈 이온에 대한 킬레이트화 부위가 4개인 NTA는 금속 이온과 상호 작용할 수 있는 부위가 3개뿐인 금속 킬레이트 정제 시스템보다 니켈을 더 단단히 결합합니다. 여분의 킬레이트화 부위는 니켈 이온 침출을 방지하여 다른 금속 킬레이트화 정제 시스템을 사용하여 얻은 것보다 결합력과 순도가 더 높은 단백질 제제를 생성합니다. QIAexpress 시스템은 baculovirus, 포유류 세포, 효모, 박테리아를 포함한 모든 발현 시스템에서 His 태그 단백질을 정제하는 데 사용할 수 있습니다.
| 변성제 | 세제 | 환원제 | 기타 | 염 | 장기 보관용 |
|---|---|---|---|---|---|
| 6 M Gu·HCl | 2% Triton X-100 | 20mM β-ME | 50% 글리세롤 | 4M MgCl2 | 최대 30% 에탄올 |
| 8 M Urea | 2% Tween 20 | 10mM DTT | 20% 에탄올 | 5mM CaCl2 | 또는 100mM NaOH |
| 1% CHAPS | 20mM TCEP | 20mM 이미다졸 | 2M NaCl |
Ni-NTA 기질은 His 태그 단백질을 신뢰할 수 있는 단일 단계로 정제하여 다음을 포함한 모든 응용 분야에 적합합니다.
| Features | Specifications |
|---|---|
| Applications | 단백질체학 |
| Tag | 6xHis tag |
| Bead size | 45~165µm |
| Special feature | 배치 및 컬럼 정제 |
| FPLC | 아니요 |
| Processing | 수동 |
| Support/matrix | Sepharose CL-6B |
| Start material | 세포 용해물 |
| Scale | 대용량 |
| Binding capacity | 최대 50mg/ml |
| Gravity flow or spin column | 중력류 |
| Yield | 100µg~100mg |