✓ 全天候自动处理在线订单
✓ 博学专业的产品和技术支持
✓ 快速可靠的(再)订购
特点
- 快速纯化高质量 DNA
- 无需有机萃取或酒精沉淀
- 稳定的高产量
- 完全去除污染物和抑制物
- 在 QIAcube Connect 上自动纯化 1–12 个样本
产品详情
QIAamp DNA Kits 可从组织、拭子、CSF、血液、体液或尿液洗涤细胞中进行基于硅胶膜技术的核酸纯化。此外,还可从少量新鲜或冷冻血液、组织和干血斑中纯化基因组和线粒体 DNA。不需要机械均质化,因为会对组织进行酶裂解。使用专用的 QIAamp DNA Mini QIAcube Kit,可在 QIAcube Connect 上自动纯化 1–12 个样本的 DNA。使用 QIAamp DNA Micro Kit 和 QIAamp DNA Mini Kit 纯化 DNA 的操作也可在 QIAcube Connect 上自动执行。
QIAamp DNA Mini 标准方案也可使用 TRACKMAN Connected 系统执行。
绩效
QIAamp DNA Micro Kit 将硅胶膜的选择性附着属性与 20 至 100 µL 的灵活洗脱体积相结合。经 QIAamp DNA Micro Kit 纯化的 DNA 不含蛋白质、核酸酶及其他杂质,适用于荧光定量 PCR(请参阅图“ 从少量样本中高效纯化 DNA”)和激光显微切割 (laser microdissection, LMD) PCR(请参阅图“ 激光显微切割 PCR”)等高灵敏度下游应用。纯化的 DNA 还可用于短串联重复 (short-tandem repeat, STR) 基因分型、单核苷酸多态性 (single-nucleotide polymorphism, SNP) 基因分型或药物基因组学研究。
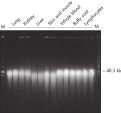
QIAamp DNA Mini Kit 使用快速离心柱或真空程序简化从组织样本中分离 DNA 的过程,所得 DNA 片段高达 50 kb(请参阅图“ 纯化高达 50 kb 的基因组 DNA”)。这种长度的 DNA 可完全变性,并具有最高扩增效率。核酸或 DNA 的产量取决于起始材料(请参阅表“使用 QIAamp DNA Mini Kit 获得的典型产量”)。使用 QIAamp DNA Mini Kit 纯化的 DNA 可用于多种下游应用,包括 PCR 和荧光定量 PCR、Southern blotting、SNP 和 STR 基因分型以及药物基因组学研究。
| 样本 | 数量 | 总核 酸产量 (µg)* | DNA 产量 (µg)† |
|---|---|---|---|
| 血液 | 200 µL | 4–12 | 4–12 |
| 血沉棕黄层 | 200 µL | 25–50 | 25–50 |
| 细胞 | 106 | 20–30 | 15–20 |
| 肝脏 | 25 mg | 60–115 | 10–30 |
| 大脑 | 25 mg | 35–60 | 15–30 |
| 肺 | 25 mg | 25–45 | 5–10 |
| 心脏 | 25 mg | 15–40 | 5–10 |
| 肾脏 | 25 mg | 40–85 | 15–30 |
| 脾脏 | 10 mg | 25–45 | 5–30 |
QIAamp DNA Mini QIAcube Kit 的 DNA 产量大小可高达 50 kb:这种长度的 DNA 可完全变性,并具有最高扩增效率。纯化的 DNA 可用于多种应用,其中包括:
- 病毒研究
- 细菌研究
- 真菌研究
- 癌症研究
- 人类基因检测研究
- 亲子鉴定
- 法证分析
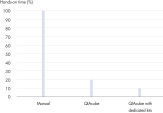
QIAamp DNA Mini QIAcube Kit 通过快速离心柱程序简化从组织样本中分离 DNA 的过程。该试剂盒包括随 QIAamp 离心柱和洗脱管预装的转子适配器,更便利且更省时(请参阅图“ 非常省时”)。
原理
在 QIAamp DNA 纯化过程中,DNA 会特异性结合至 QIAamp MinElute 或 QIAamp 硅胶膜上,而污染物则被洗除。无需酚-氯仿提取。PCR 抑制物(如二价阳离子和蛋白质)会通过两个高效的清洗步骤被完全去除,留下纯净的 DNA,然后用水或试剂盒附带的缓冲液中进行洗脱。
QIAamp DNA 技术可从人体组织样本中提取可随时用于 PCR 和印迹操作流程的基因组、线粒体、细菌、寄生虫或病毒 DNA。
QIAamp 样本制备技术已获得完全授权,因此 QIAamp 纯化核酸可用于任何分子检测或其他下游应用而不会有专利侵权风险。
程序
QIAamp DNA Mini Kit 中经过优化的缓冲液和酶可裂解样本、稳定核酸并增强 QIAamp 硅胶膜对 DNA 的选择性吸附。加入酒精,并将裂解物加到 QIAamp 离心柱上。
使用洗涤缓冲液去除杂质,然后在水或低盐缓冲液中洗脱纯净的即用型 DNA。
不需要机械均质化,因为会对组织进行酶裂解。离心柱程序很方便,这意味着实际操作时间仅需 20 分钟(裂解时间因样本来源而异)。
可使用微型离心机处理样本,如果处理的是血液或其他体液,则可使用 QIAvac 24 Plus。此外,QIAamp DNA Mini Kit 采用严格的裂解程序,令其成为从细菌或寄生虫中纯化基因组 DNA 的理想之选。
如需进一步减少手动操作时间,基因组 DNA 的纯化可在 QIAcube 上自动进行。QIAamp DNA Accessory Set A 提供在 QIAcube 上使用 12×QIAamp DNA Mini Kit (50) 分离基因组、线粒体、细菌、寄生虫或病毒 DNA 所需的额外缓冲液和试剂。
真空处理
可以用真空而不是离心来处理血液或其他体液,从而提高 DNA 纯化的速度和便利性。可使用 VacValves 和 VacConnectors 将 QIAamp Mini 离心柱安装在 QIAvac 24 Plus 歧管上。
如果样本流速差异显著,应使用 VacValves 以确保一致的真空度。使用一次性 VacConnectors 以避免任何交叉污染。如果使用 VacConnectors,则还可以通过 QIAvac Luer Adapters 在 QIAvac 6S 上执行 QIAamp 离心程序。
在 QIAcube Connect 上自动分离 DNA
专用的 QIAamp DNA Mini QIAcube Kit 配备预装在 QIAcube 仪器转子适配器中的离心柱和洗脱管,可消除因加载的转子适配器不正确而出错的风险。专用试剂盒系根据 QIAcube 的要求定制,可减少浪费。
QIAcube Connect 上的简单方案(裂解、结合、洗涤和洗脱)和自动方案只需极少的用户交互。
屡获殊荣的 QIAcube Connect 采用先进技术处理 QIAGEN 离心柱,可将自动化、低通量样本制备无缝集成到实验室工作流程中。纯化程序中的所有步骤均全自动执行 — 每次运行最多可处理 12 个样本。QIAcube Connect 和专用的 QIAamp DNA Mini QIAcube Kit 可成为快速、简易、方便地纯化 DNA 的成功组合。
应用
QIAamp DNA Micro Kit 程序适用于多种样本材料,包括少量血液、血卡、尿液、小组织样本(包括激光显微切片)。
QIAamp DNA Mini Kit 非常适合纯化最常用人体组织样本中的 DNA,包括肌肉、肝脏、心脏、大脑、骨髓和其他组织、拭子(口腔、眼、鼻、咽及其他)、CSF、血液、体液及尿液洗涤细胞。可在 20 分钟内从多达 25 mg 组织或多达 200 µl 液体中纯化 DNA,并洗脱于 50–200 µl 中。
QIAamp DNA Mini QIAcube Kit 用于在 QIAcube Connect 上自动分离基因组、线粒体、寄生虫或病毒 DNA。样本来源包括组织、口腔拭子、CSF 和尿液洗涤细胞。
| 特点 | QIAamp DNA Micro Kit | QIAamp DNA Mini Kit |
|---|---|---|
| 应用 | Real-time PCR、STR 分析、LMD-PCR | PCR、Southern blotting |
| 洗脱体积 | 20–100 µL | 50–200 µL |
| 格式 | 离心柱 | 离心柱 |
| 主要样本类型 | 全血 | 全血、组织、细胞 |
| 处理 | 手动(离心或真空) | 手动(离心或真空) |
| 总 RNA、miRNA、 poly A+ mRNA、DNA 或蛋白质的纯化 | 基因组 DNA、线粒体 DNA | 基因组 DNA、线粒体 DNA、 细菌 DNA、寄生虫 DNA、病毒 DNA |
| 样本量 | 1–100 µL | 200 µL/25 mg/5 x 106 |
| 技术 | 硅胶膜技术 | 硅胶膜技术 |
| 每次运行或每次制备的时间 | 30 分钟 | 20 分钟 |
| 产量 | <3 µg | 4–30 µg |
辅助数据和图表
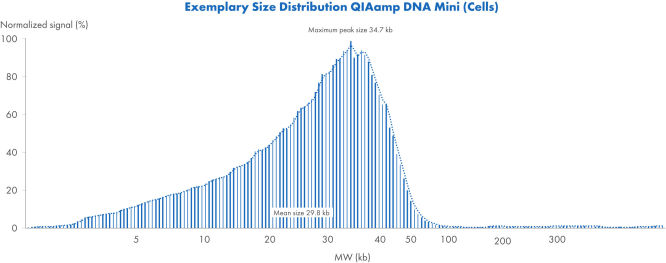
使用 QIAamp DNA Mini Kit(细胞)的典型大小分布
使用 QIAamp DNA Mini Kit 从细胞中提取 DNA 的典型大小分布。使用 Femto Pulse System 分析了样本,并将信号归一化为最大峰值。