✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 970480
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Comprehensive results in real time
- Accurate quantification of mutations in the EGFR gene
- Easy detection of complex mutations
- Sequence context provides built-in control of the assay
- Flexible post-run analysis
Product Details
The EGFR Pyro Kit is a molecular detection kit for the identification of mutations and deletions in the EGFR gene. The kit provides primers and reagents for amplification of the EGFR gene, plus buffers, primers, and reagents for detection and quantification of mutations in real time using Pyrosequencing technology on the PyroMark Q24 System.
Principle
The EGFR Pyro Kit is used for quantitative measurements of mutations in codons 719, 768, 790, 858, and 861, as well as deletions and complex mutations in exon 19 of the human EGFR gene in real time using Pyrosequencing technology on the PyroMark Q24 System. The EGFR gene encodes the epidermal growth factor receptor (EGFR) protein. Mutations in the tyrosine kinase domain of the EGFR gene can enable tumor growth and progression. EGFR mutations are found in approximately 10% of non-small cell lung cancer incidences in the US and 35% in East Asia. Additionally, EGFR mutations are found in 6% of brain tumors.
The following mutations are detected.
- Exon 18 (719): G719A, G719C, G719S
- Exon 19 (Del): 20 deletions and complex mutations
- Exon 20 (768 and 790): S768I, T790M
-
Exon 21 (858–861): L858R, L861Q, L861R
Procedure
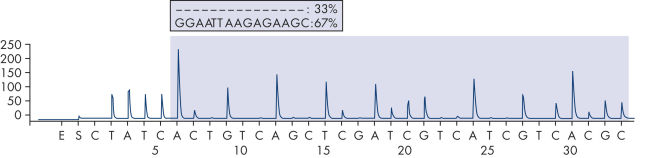
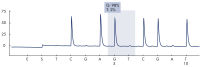
After PCR using primers targeting exons 18, 19, 20, and 21, the amplicons are immobilized on Streptavidin Sepharose High Performance beads. Single-stranded DNA is prepared, and the corresponding sequencing primers anneal to the DNA. The samples are then analyzed on the PyroMark Q24 System using a run setup file and a run file. It is recommended to use the EGFR Plug-in Report to analyze the run. This report ensures that the correct LODs are used and different sequences to analyze are automatically used to detect all mutations and deletions. However, the run can also be analyzed using the analysis tool integral to the PyroMark Q24 System (see figures " Pyrogram trace of a normal genotype in codon 719", " Pyrogram trace of a normal genotype in codon 768", " Pyrogram trace of a normal genotype in exon 19", and " Pyrogram trace of a GGAATTAAGAGAAGC deletion in exon 19"). The "Sequence to Analyze" can be then adjusted for detection different deletions in exon 19 and for rare mutations in the other exons after the run.
See figures
Applications
The EGFR Pyro Kit is used for measuring mutations in codons 719, 768, 790, 858, and 861, as well as deletions and complex mutations in exon 19 of the human EGFR gene.
The following mutations are detected.
- Exon 18 (719): G719A, G719C, G719S
- Exon 19 (Del): 20 deletions and complex mutations
- Exon 20 (768 and 790): S768I, T790M
- Exon 21 (858–861): L858R, L861Q, L861R
Supporting data and figures
Pyrogram trace of a GGAATTAAGAGAAGC deletion in exon 19.