✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
QuantiTect Virus Kit (1000)
カタログ番号 / ID. 211015
✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
特徴
- シングルおよびマルチプレックスアッセイにおける高い感度
- 同じ反応でウイルスRNAおよび/またはDNAを検出
- 弱い陽性シグナルを明確に検出
- 迅速なユニバーサル2ステッププロトコール
- より多くのサンプルインプットでより高い感度が得られる5xマスターミックス
製品詳細
パフォーマンス
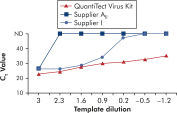
QuantiTect Virus Kitsを使用する増幅では、CT値が高い低テンプレート量でも、広範な希釈液で急峻なシグモイド曲線が得られます(図「 広いダイナミックレンジにおけるCT値の明確な決定」参照)。これにより、リアルタイムPCRでのウイルス核酸定量で、正確なCT値の決定が可能になります。
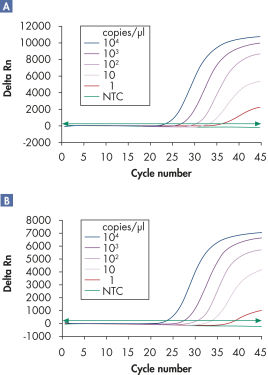
マルチプレックスアッセイでは、感度を損なうことなく、広い直線範囲にわたって、複数のウイルスRNAおよび/またはDNAターゲットと内部コントロールを検出できます(図「 広い直線範囲にわたるウイルスRNAの確実な検出」および「 少量のウイルスRNAの検出の向上」参照)。
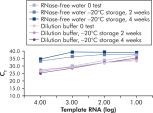
キットに付属のQuantiTect Nucleic Acid Dilution Bufferは、希釈および反応セットアップ中にRNAおよびDNAスタンダードを安定化させ、チューブやピペットチップなどのプラスチック表面での核酸の損失を防ぎます。このバッファーにより、ウイルス核酸の定量に使用されるスタンダードの信頼性の高い希釈が可能になり、低~高CT値までの広い直線範囲が得られ、変性することなく標準を長期間保存できます(図「 RNA標準の信頼性の高い希釈と保存」参照)。
原理
QuantiTect Virus Kitsは、シングルまたはマルチプレックスアッセイで、最初の試みでウイルス核酸を高感度で検出します(フローチャート「 QIAGENマルチプレックスキット」参照)。最適化されたマスターミックスは、マルチプレックス反応のPCR産物が、対応するシングル増幅反応のPCR産物と同じ効率と感度で増幅されることを保証します。
コントロール遺伝子と標的遺伝子を別々の反応ではなく、同じ反応で増幅すると、ハンドリングエラーが最小限に抑えられ、遺伝子定量の信頼性が向上します。QuantiTect Virus Bufferには、K+イオンとNH4+イオンのバランスの取れた組み合わせと、独自の合成Factor MP安定化剤が含まれており、ともにプライマーとプローブの核酸テンプレートへの安定的かつ効率的なアニーリングを促進し、高いPCR効率を実現します(図「 独自のPCRバッファー」参照)。さらに、Sensiscript Reverse Transcriptaseの独自の組成により、ウイルスRNAの高感度逆転写が保証され、HotStar Plus DNA Polymeraseは厳密なホットスタートを提供し、非特異的産物の形成を防止します。
| キットのコンポーネント | 特徴 | メリット | |
|---|---|---|---|
| 5x QuantiTect Virus Master Mix | 濃縮マスターミックス | 高濃度で、高感度ウイルス検出できるように最適化 | 感度を上げるため、より多くのテンプレート量をアッセイに添加可能 |
| HotStarTaq Plus DNA Polymerase | 95ºC 5分で活性化 | 室温でqPCR反応をセットアップ | |
| QuantiTect Virus Buffer | NH4+ イオンとK+ イオンのバランスの取れた組み合わせ | 特異的なプライマーアニーリングにより信頼性の高いPCR結果を保証 | |
| 合成Factor MP | 同じチューブで最大4つの遺伝子を高い信頼性でマルチプレックス分析 | ||
| 追加キットコンポーネント | QuantiTect Virus RT Mix | Sensiscript Reverse Transcriptaseの独自の組成を含む | ウイルスRNAを高感度検出できるように最適化 |
| QuantiTect Nucleic Acid Dilution Buffer | 核酸スタンダードの希釈および保存のための独自のバッファー組成。 | 希釈および反応セットアップ中のRNAおよびDNAスタンダードを安定化し、チューブやピペットチップなどのプラスチック表面での核酸のロスを防ぎます。 |
操作手順
QuantiTect Virus Kitsは、配列特異的プローブを使用してウイルス核酸(RNAおよび/またはDNA)および内部コントロールの高感度リアルタイムPCR分析を提供します。反応は、逆転写ステップの有無にかかわらず実施でき、RNAターゲット、DNAターゲット、またはRNAとDNAターゲットの両方を検出するマルチプレックスアッセイの柔軟な設計が可能です。迅速で信頼性の高い結果を得るためには、ハンドブックのプロトコールに従ってください。
キットは、マスターミックスにROXパッシブレファレンス色素を含むものと含まないものがあります(表参照)。
| ROX色素 | キット | 互換性のあるサイクラー |
|---|---|---|
| マスターミックスに含まれる | QuantiTect Virus Kit | Applied Biosystems 7500を除くApplied Biosystemsのすべてのサイクラー |
| 別のチューブで提供 | QuantiTect Virus +ROX Vial Kit | Applied Biosystems 7500および Bio-Rad、Cepheid、Eppendorf、QIAGEN、Roche、Agilent、およびその他のサプライヤーのサイクラー |
ウイルス検出を含む迅速かつ高感度のエンドポイント1ステップRT-PCRアプリケーションには、QIAGEN OneStep RT-PCR Kitの使用を推奨します。
アプリケーション
裏付けデータと数値
広いダイナミックレンジでCT 値を明確に決定。
仕様
| 特徴 | 仕様 |
|---|---|
| Applications | ウイルス検出 |
| SYBR Green I or sequence-specific probes | 配列特異的プローブ |
| Real-time or endpoint | リアルタイム |
| Reaction type | 逆転写およびPCR |
| Thermal cycler | ほとんどのリアルタイムサイクラー(LightCycler® 1.xおよび2.0などのキャピラリーサイクラーを除く) |
| Sample/target type | RNAおよび/またはDNAターゲット |
| With or without ROX | マスターミックス中のROXと別バイアルとしてのROXが利用可能 |
| Single or multiplex | シングルプレックスまたはマルチプレックス |