✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 28604
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- 最小限の溶出量
- 迅速な操作と容易な取り扱い
- 再現性のある高い回収率
- ゲル解析をより簡便化するGelPilot Loading Dye付き
Product Details
MinElute Gel Extraction Kitは、スピンカラム、バッファー、コレクションチューブにより構成され、シリカメンブレンによりDNAフラグメント(70 bp~4 kb)を400 mgまでのゲル切片から精製します。スピンカラムは微量(わずか10 µl)で溶出できるようにデザインされているため、高濃度のDNAが高収量で得られます。pH指示薬入りバッファーにより、DNAがスピンカラムに結合する最適なpHを簡単にチェックできます。MinEluteシステムで精製したDNAフラグメントはシークエンシング、マイクロアレイ解析、ライゲーション、トランスフォーメーション、制限酵素解析、標識反応、マイクロインジェクション、PCR、in vitro転写反応のような幅広いアプリケーションに即使用できます。
Performance
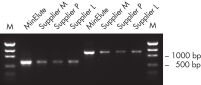
MinElute Gel Extraction Kit には、ヌクレオチド、酵素、塩類、アガロース、臭化エチジウムなどの夾雑物をDNAサンプルから除去するため、様々なダウンストリームアプリケーションに適した高濃度なDNAを提供します(図"高濃度なDNA精製")。
MinElute QIAquick Gel Extraction Kitは、ゲル抽出用のスピンカラムが含まれています。マイクロ遠心機または吸引マニホールドを使用することにより、70 bp~4 kbの高濃度なDNAが迅速に精製されます(4~10kbのDNAフラグメントの精製にはQIAquick Gel Extraction Kitを、70 bp以下あるいは10 kb以上のフラグメントにはQIAEX II Gel Extraction System をご利用ください)。
Principle
MinElute PCR Purification Kitでは、高塩濃度バッファーによりDNAを結合し、低塩濃度バッファーあるいは水によりDNAを溶出するシリカゲルメンブレン を利用しています。シリカメンブレンテクノロジーにより樹脂漏れ、および懸濁液関連の問題や不便さが解消されます。それぞれの用途に応じて至適化された結合バッファーにより、種々のサイズのDNAフラグメントを選択的に吸着します。
Gel Loading Dye
より迅速で簡便なサンプル処理と解析を実現するため、ゲル電気泳動用のローディング色素が付いています。GelPilot Loading Dye は3 種類のマーカー色素(xylene cyanol、bromophenol blueおよびorange G)が入っており、短いDNAフラグメントのゲルからの流出を回避でき、アガロースゲルの泳動時間の至適化が簡単に行なえます(図 " GelPilot Loading Dye")。
See figures
Procedure
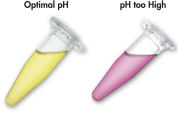
MinEluteシステムでは結合-洗浄-溶出という簡単な操作だけでDNAのクリーンアップが可能です(フローチャート" MinElute操作手順"参照)。ゲル切片をpH指示薬を含んだバッファーに溶解させると、DNA結合の至適pHを容易に決めることができます(図 "pH 指示薬")。DNAを含むバッファーをMinElute Spin Column にアプライします。DNAは添付のバッファー中の高塩濃度の条件下でシリカゲルメンブレンに吸着します。不純物は洗い流され、純粋なDNAを添付の低塩濃度溶出バッファー、あるいは水で溶出します。得られたDNAは、その後のアプリケーションにも即使用可能です。
操作法
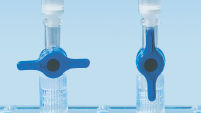
MinElute Spin Column は2つの簡便な操作法オプションのためにデザインされました(図" MinElute操作手順")。スピンカラムは簡便な卓上型マイクロ遠心機あるいはQIAvac 24 Plus のようなルアーコネクター付き吸引装置で使用できます。MinElute Gel Extraction Kitは他のQIAGENスピンカラムを利用したキット同様にQIAcubeでの完全自動化が可能で、 標準化された再現性の高い結果が得られます(図 "Spin Column操作オプション A、 B、 C、 D、およびE")。
See figures
Applications
MinEluteあるいはQIAquickシステムで精製したDNAフラグメントは以下のようなアプリケーションに即使用可能です。
- 次世代シークエンシングを含むシークエンシング
- マイクロアレイ解析
- ライゲーションやトランスフォーメーション
- 制限酵素分解
- 標識反応
Supporting data and figures
Spin column handling options — C.