QuantiTect Reverse Transcription Kit
For fast cDNA synthesis enabling sensitive real-time two-step RT-PCR for gene expression analysis
For fast cDNA synthesis enabling sensitive real-time two-step RT-PCR for gene expression analysis
Cat. No. / ID: 205311
Using the QuantiTect Reverse Transcription Kit, contaminating genomic DNA in RNA samples is effectively and rapidly removed with the unique gDNA Wipeout Buffer (see figure Effective genomic DNA removal for accurate real-time RT-PCR). Elimination of genomic DNA is crucial for accurate gene expression results, and design of RNA-specific primers or probes is not always possible. With gDNA Wipeout Buffer, time is saved and costs are reduced, since a separate DNase digestion is unnecessary, either during or after purification of RNA samples.
The high RNA affinity of Quantiscript Reverse Transcriptase, in combination with Quantiscript RT Buffer, enables high yields of cDNA from any RNA template (see table “Higher cDNA yields for less abundant transcripts”). Even difficult templates, such as those with high GC-content or complex secondary structure, are successfully reverse transcribed.
| CT values for IL12A (low expression) |
CT values for IL1RN (higher expression) |
|||
|---|---|---|---|---|
| Input RNA (ng) | QIAGEN | Supplier AII | QIAGEN | Supplier AII |
| 1000 | 30.9 | 32.0 | 23.1 | 24.9 |
| 100 | 34.2 | 35.4 | 26.3 | 26.6 |
| 10 | 37.8 | 46.8 | 29.7 | 30.3 |
| 1 | N.D. | N.D. | 32.4 | 34.5 |
The RT Primer Mix contains a specially optimized mix of oligo-dT and random primers that enable cDNA synthesis from all regions of RNA transcripts, even from 5' regions (see figure Sensitive detection of a target at the 5' region of a 12.5 kb transcript). In contrast to kits from other suppliers, the QuantiTect Reverse Transcription Kit provides high yields of cDNA template for real-time PCR analysis regardless of where the amplified target region is located on the transcript, and provides greater sensitivity in the detection of low-abundance genes (see figure " Higher sensitivity in real-time, two-step RT-PCR"). The QuantiTect Reverse Transcription Kit also enables greater reproducibility in real-time RT-PCR.
QuantiTect Reverse Transcriptase is a novel blend of Omniscript and Sensiscript Reverse Transcriptases, which has a high affinity for RNA and is capable of cDNA synthesis from a wide range of RNA amounts (10 pg to 1 µg). In contrast to kits from other suppliers, the QuantiTect Reverse Transcription Kit provides high yields of cDNA template for real-time PCR analysis regardless of where the amplified target region is located on the transcript. Even difficult templates, such as those with high GC-content or complex secondary structure, are successfully reverse transcribed. QuantiTect RT Buffer has also been optimized to be compatible with real-time PCR buffer.
To obtain accurate results in real-time RT-PCR gene expression assays, it is important that only cDNA is amplified and detected. Interference by genomic DNA can be avoided by designing primers or probes that span an exon/exon boundary. However, in cases where this is not possible (e.g., the cDNA is from a single-exon gene), it is essential that the starting RNA sample is free of genomic DNA. Using the QuantiTect Reverse Transcription Kit, contaminating genomic DNA in RNA samples is effectively and rapidly removed with unique gDNA Wipeout Buffer. Time is saved and costs are reduced, since a separate DNase digestion not required, either during or after purification of RNA samples. Also, design of RNA-specific primers or probes is unnecessary.
| Component | Benefits |
|---|---|
| gDNA Wipeout Buffer | Detection of RNA only in real-time RT-PCR |
| Quantiscript Reverse Transcriptase | Use of a wide range of RNA amounts (10 pg to 1 µg RNA) High sensitivity |
| Quantiscript RT Buffer | Read-through of difficult templates |
| RT Primer Mix | cDNA synthesis from all regions of transcripts, even from 5' regions |
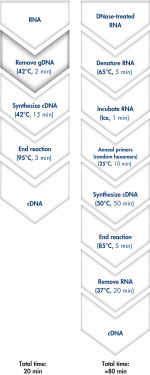
Genomic DNA removal and cDNA synthesis take only 20 minutes with the QuantiTect Reverse Transcription Kit (see flowchart " Fast and convenient cDNA synthesis"). The procedure is fast and convenient, since both reactions are run using the same incubation temperature and are set up using master mixes.
The QuantiTect Reverse Transcription Kit includes everything you need for fast cDNA synthesis. Purified RNA is briefly incubated in gDNA Wipeout Buffer to effectively remove contaminating genomic DNA. In contrast to other methods, the RNA sample is then used directly in reverse transcription, using a master mix prepared from Quantiscript Reverse Transcriptase, Quantiscript RT Buffer, and RT Primer Mix. With Quantiscript Reverse Transcriptase, RNA can be transcribed at low temperatures, even through complex 2° structure, ensuring that the RNA will stay intact — the entire reaction takes place at 42°C and is then inactivated at 95°C. Additional steps for RNA denaturation, primer annealing, and RNase H digestion are not necessary.
The QuantiTect Reverse Transcription Kit allows highly efficient and sensitive real-time RT-PCR for all types of starting material, including laser-microdissected samples and tissue biopsies.
Genomic DNA removal and cDNA synthesis take only 20 minutes with the QuantiTect Reverse Transcription Kit. The procedure is fast and convenient since both reactions are run using the same incubation temperature and are set up using master mixes. In contrast, the procedure for the kit from Supplier I is much longer and requires more "hands-on time" due to additional pipetting steps and frequent changes in incubation temperature.
| Features | Specifications |
|---|---|
| Applications | Quantification of (even low-abundance) transcripts |
| Sample/target type | RNA template |
| Enzyme activity | Reverse transcription |
| Real-time or endpoint | Real time |
| Reaction type | Two-step, cDNA production, genomic DNA digestion |
| Single or multiplex | Single |
| Mastermix | No |