✓ Procesamiento automático sin interrupción de pedidos en línea
✓ Servicio técnico y para productos experto y profesional
✓ Realización y repetición de pedidos rápidas y fiables
QIAamp DNA Micro Kit (50)
N.º de cat. / ID. 56304
✓ Procesamiento automático sin interrupción de pedidos en línea
✓ Servicio técnico y para productos experto y profesional
✓ Realización y repetición de pedidos rápidas y fiables
Características
- Purificación rápida de ADN de gran calidad
- Sin extracción orgánica ni precipitación con alcohol
- Rendimiento alto y constante
- Eliminación completa de contaminantes e inhibidores
- Purificación automatizada de entre 1 y 12 muestras en QIAcube Connect
Detalles del producto
Los QIAamp DNA Kits permiten la purificación de ácidos nucleicos basada en membranas de gel de sílice a partir de tejido, frotis en hisopos, líquido cefalorraquídeo, sangre, líquidos corporales o células lavadas de la orina. Además, el ADN genómico y mitocondrial puede purificarse a partir de cantidades pequeñas de sangre fresca o congelada, tejido y gotas de sangre seca. No es necesario aplicar homogeneización mecánica, ya que los tejidos se lisan de forma enzimática. La purificación de ADN de entre 1 y 12 muestras puede automatizarse en QIAcube Connect gracias al QIAamp DNA Mini QIAcube Kit. La purificación de ADN con el QIAamp DNA Micro Kit y el QIAamp DNA Mini Kit también puede automatizarse en el instrumento QIAcube Connect.
Los protocolos estándar de QIAamp DNA Mini también pueden llevarse a cabo mediante el uso del sistema TRACKMAN Connected.
Rendimiento
El QIAamp DNA Micro Kit combina las propiedades de unión selectiva de una membrana de gel de sílice con volúmenes de elución flexibles de entre 20 y 100 µl. El ADN que se purifica con el QIAamp DNA Micro Kit no contiene proteínas, nucleasas ni otras impurezas, y es apto para su uso en aplicaciones anterógradas, como real-time PCR (consulte la figura “ Purificación eficaz de ADN a partir de muestras de tamaño pequeño”) y PCR de microdisección láser (LMD-PCR) (consulte la figura “ PCR de microdisección láser”). El ADN purificado también puede utilizarse en procesos de genotipado de repeticiones cortas en tándem (STR), genotipado de polimorfismos de un solo nucleótido (SNP) o investigación farmacogenómica.
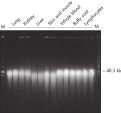
El QIAamp DNA Mini Kit simplifica el aislamiento de ADN a partir de muestras de tejido gracias al uso de procedimientos de columnas de centrifugación rápidas o métodos de vacío, lo que permite obtener ADN de hasta 50 kb (consulte la figura “ Purificación de ADN genómico de hasta 50 kb”). El ADN de esta longitud se desnaturaliza por completo y ofrece la mayor eficacia de amplificación. Los rendimientos de ácido nucleico o ADN dependen del material inicial (consulte la tabla “Rendimientos habituales con QIAamp DNA Mini Kit”). El ADN purificado a partir del QIAamp DNA Mini Kit puede utilizarse en una amplia gama de aplicaciones anterógradas, como PCR, real-time PCR cuantitativo, Southern blotting, genotipado SRT y SNP e investigación farmacogenómica.
| Muestra | Cantidad | Rendimientos de ácido nucleico total (µg)* | Rendimientos de ADN (µg)† |
|---|---|---|---|
| Sangre | 200 µl | 4-12 | 4-12 |
| Capa leucoplaquetaria | 200 µl | 25-50 | 25-50 |
| Células | 106 | 20-30 | 15-20 |
| Hígado | 25 mg | 60-115 | 10-30 |
| Cerebro | 25 mg | 35-60 | 15-30 |
| Pulmón | 25 mg | 25-45 | 5-10 |
| Corazón | 25 mg | 15-40 | 5-10 |
| Riñón | 25 mg | 40-85 | 15-30 |
| Bazo | 10 mg | 25-45 | 5-30 |
El QIAamp DNA Mini QIAcube Kit obtiene ADN de hasta 50 kb: El ADN de esta longitud se desnaturaliza por completo y ofrece la mayor eficacia de amplificación. El ADN purificado puede usarse en diversas aplicaciones, como las siguientes:
- Investigación sobre virus
- Investigación sobre bacterias
- Investigación sobre hongos
- Investigación sobre el cáncer
- Investigación en pruebas de genética humana
- Pruebas de paternidad
- Análisis forenses
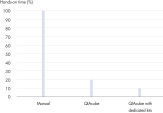
El QIAamp DNA Mini QIAcube Kit simplifica el aislamiento de ADN a partir de muestras de tejido gracias al uso de procedimientos de columnas de centrifugación rápidas. El kit incluye adaptadores de rotor precargados con tubos de elución y columnas de centrifugación de QIAamp, lo que proporciona un procedimiento más práctico y que permite ahorrar tiempo (consulte la figura “ Ahorro de tiempo significativo”).
Principio
Durante el procedimiento de purificación de ADN con QIAamp, el ADN se une de forma específica a la membrana de gel de sílice de QIAamp MinElute o QIAamp, mientras los contaminantes la atraviesan. No se requiere extracción con fenol-cloroformo. Los inhibidores de la PCR, como los cationes divalentes y las proteínas, se eliminan por completo en dos eficientes pasos de lavado, dejando el ADN puro para su elución en agua o en un tampón incluido en el kit.
La tecnología de procesamiento de ADN de QIAamp permite obtener ADN genómico, mitocondrial, bacteriano, parásito o vírico a partir de muestras de tejido humano listas para su uso en procedimientos de PCR y blotting.
La tecnología de preparación de las muestras de QIAamp cuenta con licencia completa, lo que permite utilizar los ácidos nucleicos purificados con QIAamp en cualquier ensayo molecular o cualquier aplicación anterógrada sin que exista el riesgo de incumplir la patente.
Procedimiento
Los tampones optimizados y las enzimas del QIAamp DNA Mini Kit lisan las muestras, estabilizan los ácidos nucleicos y mejoran la adsorción selectiva de ADN en la membrana QIAamp. Se añade alcohol y se cargan los lisados en la QIAamp Spin Column.
Los tampones de lavado se usan para eliminar impurezas y, a continuación, se eluye ADN puro listo para su uso en agua o en tampón bajo en sal.
No se requiere la homogeneización mecánica, ya que los tejidos se lisan de forma enzimática. Gracias al práctico procedimiento con columnas de centrifugación, el tiempo de preparación es de solo 20 minutos (los tiempos de lisis pueden variar en función del origen de la muestra).
Las muestras se pueden procesar con una microcentrifugadora o, si se va a procesar sangre u otros fluidos corporales, con QIAvac 24 Plus. Además, el riguroso proceso de lisis empleado hace que el QIAamp DNA Mini Kit sea ideal para la purificación de ADN genómico de bacterias o parásitos.
Para reducir aún más el tiempo de manipulación, la purificación del ADN genómico puede automatizarse en QIAcube. El QIAamp DNA Accessory Set A proporciona los tampones y reactivos adicionales necesarios para el aislamiento de ADN genómico, mitocondrial, bacteriano, parásito o vírico para el uso de QIAcube con 12 × QIAamp DNA Mini Kits (50).
Procesamiento de sistemas de vacío
La sangre y otros líquidos corporales pueden procesarse mediante sistemas de vacío en lugar de centrifugación, lo que permite una mayor velocidad y practicidad en la purificación de ADN. Las QIAamp Mini Spin Columns se colocan en el colector QIAvac 24 Plus con la ayuda de VacValves y VacConnectors.
Las VacValves deben utilizarse si el flujo de las muestras difiere significativamente para conseguir un vacío uniforme. Se utilizan VacConnectors desechables para evitar cualquier posible contaminación cruzada. El uso de VacConnectors también permite que los procedimientos de centrifugación de QIAamp puedan llevarse a cabo en QIAvac 6S con QIAvac Luer Adapters.
Aislamiento de ADN automatizado en QIAcube Connect
El QIAamp DNA Mini QIAcube Kit está diseñado de manera que viene precargado con columnas de centrifugación y tubos de elución en los adaptadores de rotor para los instrumentos de QIAcube, lo que elimina el riesgo de errores debido a la carga incorrecta de adaptadores de rotor. El kit especializado está diseñado para adaptarse a los requisitos de QIAcube y genera menos residuos.
Un protocolo sencillo (lisis, unión, lavado y elución) y un protocolo automatizado en QIAcube Connect que exige una interacción mínima por parte del usuario.
El galardonado instrumento QIAcube Connect utiliza la tecnología avanzada para procesar las columnas de centrifugación de QIAGEN, lo que permite una integración perfecta de la preparación automatizada de muestras de bajo volumen en el flujo de trabajo del laboratorio. Todos los pasos del procedimiento de purificación están totalmente automatizados (se pueden procesar hasta 12 muestras por serie). Cuando se utiliza QIAcube Connect y el QIAamp DNA Mini QIAcube Kit, se crea una combinación óptima que proporciona una purificación de ADN rápida, práctica y sencilla.
Aplicaciones
El procedimiento del QIAamp DNA Micro Kit es apto para una amplia gama de materiales de muestra, como pequeñas cantidades de muestras de sangre, tarjetas de sangre, orina y muestras de tejido de tamaño pequeño, como microdisecciones láser.
El QIAamp DNA Mini Kit es idóneo para la purificación de ADN de las muestras de tejido humano que suelen utilizarse habitualmente, como músculo, hígado, corazón, cerebro, médula ósea y otros tejidos, frotis de hisopos (bucal, conjuntival, nasal, faríngeo, etc.), líquido cefalorraquídeo, sangre, líquidos corporales y células lavadas de la orina. El ADN puede purificarse a partir de hasta 25 mg de tejido o 200 µl de líquido en 20 minutos, y eluirse en 50-200 µl.
El QIAamp DNA Mini QIAcube Kit es adecuado para el aislamiento automatizado de ADN genómico, mitocondrial, parásito o vírico en QIAcube Connect. Los orígenes de las muestras incluyen tejidos, hisopos de frotis bucales, líquido cefalorraquídeo y células lavadas de la orina.
| Características | QIAamp DNA Micro Kit | QIAamp DNA Mini Kit |
|---|---|---|
| Aplicaciones | Real-time PCR, análisis de STR, LMD-PCR | PCR, Southern blotting |
| Volumen de elución | 20-100 µl | 50-200 µl |
| Formato | Columna de centrifugación | Columna de centrifugación |
| Tipo de muestra principal | Sangre total | Sangre total, tejido, células |
| Procesamiento | Manual (centrifugación o vacío) | Manual (centrifugación o vacío) |
| Purificación de ARN total, miARN, ARNm poli A+, ADN o proteína | ADN genómico, ADN mitocondrial | ADN genómico, ADN mitocondrial, ADN bacteriano, ADN parásito, ADN vírico |
| Cantidad de muestra | 1-100 µl | 200 µl/25 mg/5 × 106 |
| Tecnología | Tecnología de sílice | Tecnología de sílice |
| Tiempo por serie o por preparación | 30 minutos | 20 minutos |
| Rendimiento | <3 µg | 4-30 µg |
Datos y cifras de respaldo
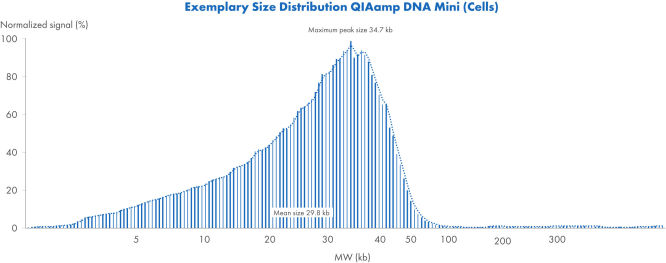
Ejemplo de distribución de tamaño con el QIAamp DNA Mini Kit (células)
Ejemplo de distribución de tamaño de ADN a partir de células con el QIAamp DNA Mini Kit. Se analizaron muestras con el Femto Pulse System y se normalizaron las señales hasta el pico máximo.