✓ Automatische Verarbeitung von Online-Bestellungen 24/7
✓ Sachkundiger und professioneller technischer und Produkt-Support
✓ Schnelle und zuverlässige (Nach-)Bestellung
Cat. No. / ID: 31314
✓ Automatische Verarbeitung von Online-Bestellungen 24/7
✓ Sachkundiger und professioneller technischer und Produkt-Support
✓ Schnelle und zuverlässige (Nach-)Bestellung
Eigenschaften
- Bis zu 300 μg Protein mit His-tag pro Säule in nur 15 Minuten
- Aufreinigung unter nativen und denaturierenden Bedingungen
- Bis zu 95 % Homogenität in einem Schritt
- Gebrauchsfertige Spin-Säulen für die schnelle automatische oder manuelle Verarbeitung
Angaben zum Produkt
Ni-NTA-Silica kombiniert Ni-NTA mit einem makroporösen Siliziumdioxid-Trägermaterial, das zur Vermeidung unspezifischer hydrophober Wechselwirkungen optimiert wurde. Ni-NTA Spin Columns (Spin-Säulen zur Aufreinigung von His-Proteinen), die im Ni-NTA Spin Kit enthalten und auch separat erhältlich sind, bieten Ni-NTA-Silica in einem praktischen Mikrospin-Format für die einfache und parallele Präparation mehrerer Proben. Sie bieten ein einfaches Verfahren zum funktionellen Screening von gentechnisch hergestellten Proteinen, zur Selektion von Klonen, die Translationsprodukte in voller Länge exprimieren, und zum Vergleich von Expressionsniveaus. Jede Spin-Säule kann bis zu 300 µg Protein mit His-tag aufreinigen. Wie alle Ni-NTA-Matrizes können Ni-NTA-Spin-Säulen für die Ein-Schritt-Proteinaufreinigung unter nativen und denaturierenden Bedingungen genutzt werden. Das Ni-NTA Spin Kit ist ein Komplettkit für die Spin-Säulen-Aufreinigung von Proteinen mit His-tag. Es kann auf dem QIAcube Connect automatisiert werden (siehe Abbildung „QIAcube Connect“).
Leistung
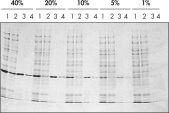
Ni-NTA Spin Columns (Spin-Säulen zur Aufreinigung von His-Proteinen), die ebenfalls im Ni-NTA Spin Kit enthalten sind, ermöglichen eine reproduzierbare, schnelle und automatisierte Aufreinigung (siehe Abbildung “Reproduzierbare, automatisierte Aufreinigung”) bei unterschiedlichen Expressionsniveaus (siehe Abbildung “Aufreinigung bei unterschiedlichen Expressionsniveaus”).
Abbildungen ansehen
Prinzip
Das QIAexpress Ni-NTA-System zur Proteinaufreinigung, einschließlich der Ni-NTA Spin Columns und des Ni-NTA Spin Kits, basiert auf der bemerkenswerten Selektivität des patentierten Nickel-Nitrilotriessigsäure(Ni-NTA)-Harzes für Proteine, die einen Affinitäts-Tag mit sechs oder mehr Histidinresten – einen His-tag – aufweisen. Diese Technologie ermöglicht eine Ein-Schritt-Aufreinigung fast aller Proteine mit His-tag aus jedem Expressionssystem unter nativen oder denaturierenden Bedingungen. NTA, das über vier Chelation-Bindestellen für Nickelionen verfügt, bindet Nickel fester als metallchelatbildende Aufreinigungssysteme, die nur drei verfügbare Bindestellen für die Interaktion mit Metallionen besitzen. Die zusätzliche Chelation-Bindestelle verhindert das Auswaschen von Nickelionen und führt zu einer höheren Bindungskapazität und damit zu Proteinpräparationen mit höherer Reinheit verglichen mit denen, die mit anderen metallchelatbildenden Aufreinigungssystemen gewonnen wurden. Das QIAexpress System kann zur Aufreinigung von Proteinen mit His-tag aus allen Expressionssystemen, einschließlich Baculovirus, Säugerzellen, Hefen und Bakterien, genutzt werden.
Verfahren
Die Aufreinigung von Proteinen mit His-tag besteht aus 4 Phasen: Zelllyse, Bindung, Waschen und Elution (siehe Abbildung “Ni-NTA-Spin-Säulen-Aufreinigung mit dem Ni-NTA-Proteinaufreinigungssystem”). Die Aufreinigung rekombinanter Proteine mit dem QIAexpress System ist nicht von der dreidimensionalen Struktur des Proteins oder des His-tags abhängig. Dadurch ist eine Ein-Schritt-Proteinaufreinigung sowohl unter nativen als auch unter denaturierenden Bedingungen, ausgehend von verdünnten Lösungen und Rohlysaten, möglich. Bis zu 600 μl eines Zelllysats werden in eine Ni-NTA-Spin-Säule geladen. Ein schneller zweiminütiger Spin bindet das markierte Protein an das Ni-NTA-Silica, während die meisten nicht markierten Proteine durchlaufen. Nach einem Waschschritt wird das aufgereinigte Protein unter milden Bedingungen (z. B. Absenkung des pH-Wertes auf 5,9 oder Zugabe von 100–500 mM Imidazol) in einem Volumen von 100–300 μl eluiert. In der Regel muss der His-tag nicht entfernt werden, da er klein und meist nicht immunogen ist. Das aufgereinigte Protein ist sofort einsatzbereit. Proteine können aus mehreren kleinen Expressionskulturen in etwa 30 Minuten (manuelles Verfahren) oder in etwa 60 Minuten (automatisiert mit QIAcube Connect) aufgereinigt werden. Starke denaturierende Agenzien und Detergenzien können für eine effiziente Solubilisierung und Aufreinigung von Rezeptoren, Membranproteinen und Proteinen, die Einschlusskörper (inclusion bodies) bilden, eingesetzt werden. Reagenzien, die eine effiziente Entfernung unspezifisch bindender Kontaminanten ermöglichen, können den Waschpuffern hinzugefügt werden (siehe Tabelle). Die aufgereinigten Proteine werden unter milden Bedingungen durch Zugabe von 100–250 mM Imidazol als Kompetitor oder durch Absenkung des pH-Werts eluiert.
Mit der Ni-NTA–His-Wechselwirkung kompatible Reagenzien:
- 6 M Guanidin-HCl
- 8 M Harnstoff
- 2 % Triton X-100
- 2 % Tween 20
- 1 % CHAPS
- 20 mM β-ME
- 10 mM DTT
- 50 % Glycerol
- 20 % Ethanol
- 2 M NaCl
- 4 M MgCl2
- 5 mM CaCl2
- ≤20 mM Imidazol
- 20 mM TCEP
Abbildungen ansehen
Anwendungen
Das QIAexpress Ni-NTA-System zur Proteinaufreinigung, einschließlich der Ni-NTA Spin Columns und des Ni-NTA Spin Kits, ermöglicht eine zuverlässige Ein-Schritt-Aufreinigung von Proteinen, die sich für jede Anwendung eignen wie:
- Struktur- und Funktionsuntersuchungen
- Kristallisierung zur Bestimmung der dreidimensionalen Struktur
- Protein-Protein- und Protein-DNA-Interaktionsassays
- Immunisierung zur Antikörperproduktion
| Eigenschaften | Ni-NTA Spin Columns | Ni-NTA Spin Kit |
| Anwendungen | Proteomik | Proteomik |
| Bead-Größe | 16–24 µm | 16–24 µm |
| Bindekapazität | Bis zu 300 µg je Spin-Säule | Bis zu 300 µg je Spin-Säule |
| Gravity-flow oder Spin-Säule | Spin-Säule | Spin-Säule |
| Verarbeitung | Automatisiert/manuell | Automatisiert |
| Maßstab | Kleiner Maßstab | Kleiner Maßstab |
| Besondere Eigenschaften | Screening mit niedrigem Durchsatz | Bis zu 95 % Homogenität in einem Schritt |
| Ausgangsmaterial | Zelllysat | Zelllysat |
| Träger/Matrix | Makroporöses Siliziumdioxid (Silica) | Makroporöses Siliziumdioxid (Silica) |
| Tag | 6xHis-tag | 6xHis-tag |
Ergänzende Daten und Abbildungen
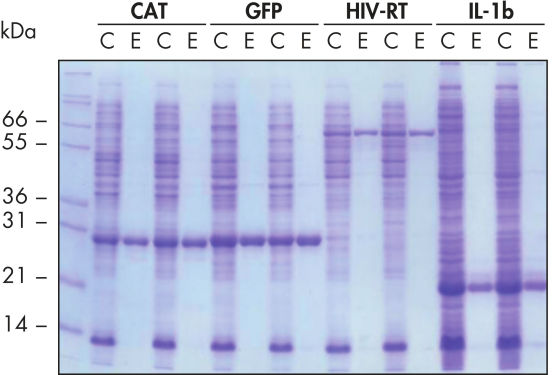
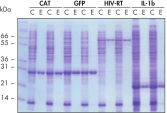
Reproduzierbare, automatisierte Aufreinigung.
Die Aufreinigung der angegebenen Proteine erfolgte in Duplikaten unter nativen Bedingungen mit Ni-NTA Spin Columns – entweder manuell oder mit einem automatisierten Verfahren auf dem QIAcube – aus geklärten E. coli-Zelllysaten, die aus 5 ml LB-Kulturen gewonnen wurden. CAT: Chloramphenicol-Acetyl-Transferase; GFP: grün fluoreszierendes Protein; HIV-RT: Reverse Transkriptase des Humanen Immundefizienz-Virus; IL-1b: Interleukin-1 beta. M: Marker; C: geklärtes Lysat (2 μl je Spur); E: Elutionsfraktion (3 μl je Spur).