✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 47017
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Lyse bacteria and fungi efficiently from soil, stool and gut samples
- Scale up to a 96 well format with the same proven technology found in the DNeasy PowerSoil Pro Kit
- Recover inhibitor-free DNA, ready to use directly in downstream NGS applications
- Achieve unbiased results that accurately represent sample alpha and beta diversity
Product Details
Extracting microbial DNA from large amounts of soil and stool samples can be challenging. QIAGEN’s new DNeasy 96 PowerSoil Pro Kit uses our innovative Pro technology, increasing efficiency of the lysis process to enable isolation of high yields of pure microbial DNA from all soil types, including compost, clay and top soil, as well as stool and gut samples. This is a 96 well format kit for manual high-throughput purification.
The DNeasy 96 PowerSoil Pro Kit is intended for molecular biology applications. This product is not intended for the diagnosis, prevention, or treatment of a disease.
Performance
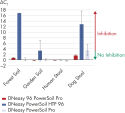
QIAGEN’s Pro technology uses new PowerBead Pro Plates for homogenization. The improved bead beating method and optimized chemistry enable more efficient lysis of bacteria and fungi. The kit also contains streamlined Inhibitor Removal Technology (IRT) to eliminate the challenging inhibitors commonly found in stool (heme, bilirubin, bile) and soil (humic/ fulvic acids) samples in even less time.
Principle
Environmental or human samples are broken down and homogenized. Cells are lysed by both mechanical and chemical means and the DNA is captured on a 96-well silica membrane in a spin plate format, washed and eluted. The DNA is then ready for downstream applications such as qPCR or next-generation sequencing.
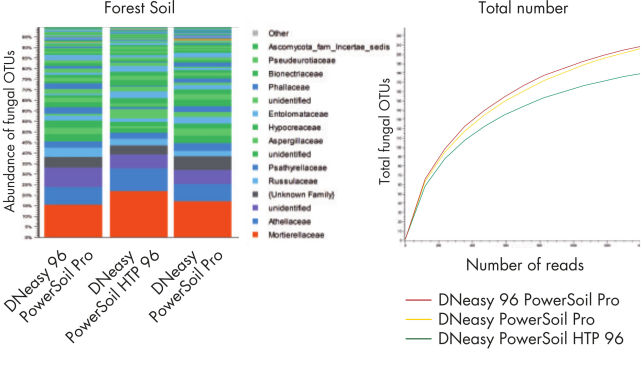
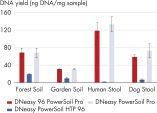
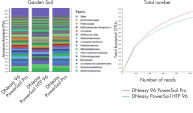
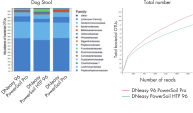
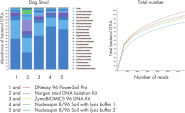
Sequencing results reveal higher alpha diversity as measured by observed operational taxonomic units (OTUs) compared to other tested methods (See figure Significantly increased DNA yields and better quality compared to other suppliers while offering comparable yields to other QIAGEN products.).
DNA isolated using the DNeasy 96 PowerSoil Pro identified more OTUs for both bacteria (See figure Increased bacterial representation.) and fungi (See figure Increased fungal representation.) compared with the legacy DNeasy PowerSoil HTP 96.
Supporting data and figures
Increased fungal representation