✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 205411
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Eliminate artifacts using the gDNA Removal Mix
- Monitor successful cDNA synthesis using the internal control
- Synthesize cDNA over a wide linear range with high-affinity transcriptase
Product Details
Note: The QuantiNova Reverse Transcription Kit (50) 205411 and (200) 205413 are temporarily unavailable. We recommend using the QuantiTect Reverse Transcription Kit (50) 205311, (200) 205313 or (400) 205314. It is fully compatible with all QuantiNova Reverse Transcription Kit applications, including two-step RT-qPCR using QuantiNova PCR kits and/or QuantiNova LNA assays or panels. It also offers integrated genomic DNA removal, ultrafast 20-minute protocol, random primers and high transcription efficiency. However, the total RNA input is limited to 1 µg, and this kit does not provide an Internal Control RNA.
Find out more about the QuantiTect Reverse Transcription Kit. If you need further assistance or have any questions about the change, please do not hesitate to contact our team of experts via your local QIAGEN representatives.
The QuantiNova Reverse Transcription Kit provides a fast and convenient procedure for cDNA synthesis with integrated genomic DNA removal and an additional internal control. Genomic DNA contamination in RNA samples is effectively eliminated by the gDNA Removal Mix. The QuantiNova Reverse Transcription Kit provides everything you need for fast and efficient reverse transcription. The synthesized cDNA is optimized for use in real-time PCR, allowing reliable quantification of targets from all regions of mRNA transcripts.
The internal control RNA can be used to test successful reverse transcription and amplification using the QuantiNova IC Probe Assay (Max label, cat. no. 205813) or QuantiNova IC Probe Assay Red 650 (Cy5 analog label, cat. no. 205824) for probe based detection. For SYBR Green based detection please order via our GeneGlobe platform: QuantiNova IC SYBR Green Assay (Available under the name HS_QIC_2467742 QuantiNova LNA PCR Reference Assay and the GeneGlobe ID: SBH1218551).
Performance
The newly developed internal control is a defined transcript (RNA molecule) that can simply be added to your samples and transcribed into cDNA. It is intended to report instrument or chemistry failures, errors in assay setup and the presence of inhibitors. The expected range of Cq (CT) values can be used to test successful reverse transcription/amplification and the reproducibility of your results.
The high RNA affinity of the QuantiNova Reverse Transcriptase enables high yields of cDNA from any RNA template. Difficult templates, such as those with high GC-content or complex secondary structure, are also successfully reverse transcribed. The Reverse Transcription Mix contains a specially optimized mix of oligo-dT and random primers that enable cDNA synthesis from all regions of RNA transcripts, including 5' regions. The kit provides high yields of cDNA template for real-time PCR analysis, regardless of where the amplified target region is located on the transcript, and provides greater sensitivity in the detection of low-abundance genes.
The QuantiNova Reverse Transcription Kit offers accurate results over a wide range of input amounts, including up to 5 µg RNA in the standard protocol, which can be doubled to accommodate exceptionally large amounts of RNA.
Principle
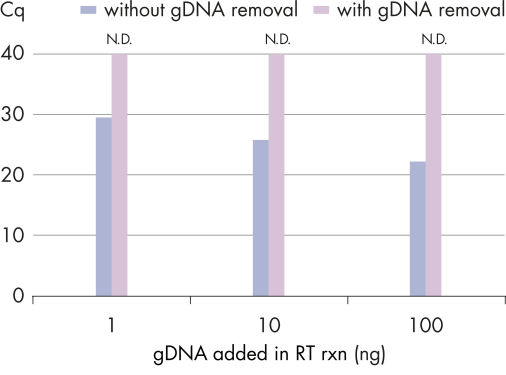
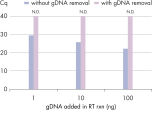
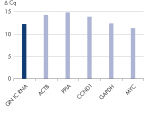
To obtain accurate results in real-time RT-PCR gene expression assays, it is important that only cDNA is amplified and detected. Interference by genomic DNA can effectively and rapidly be removed using the gDNA Removal Mix (see figure “ Efficient removal of contaminating gDNA ensures precise quantification of transcripts”). Time is saved and costs are reduced, since a separate DNase digestion during or after purification of RNA samples is not required. Also, designing RNA-specific primers or probes is not necessary.
Detecting variations in cDNA synthesis allows you to check the reproducibility of your results. The newly developed internal control is a defined transcript (RNA molecule) that can be optionally added to your samples and transcribed into cDNA. It is intended to report instrument or chemistry failures, errors in assay setup and the presence of inhibitors. Because the internal control behaves comparable to real transcripts, it can be used to confirm successful reverse transcription and amplification in subsequent qPCR (see figure “ Internal control reliably detects presence of inhibitors”).
| Component | Benefits |
|---|---|
| gDNA Removal Mix | Detection of RNA only in real-time RT-PCR |
| Internal Control RNA | Verification of successful RT-PCR performance |
| QuantiNova Reverse Transcriptase | Use of a wide range of RNA amounts (10 pg – 5 μg RNA) High sensitivity |
| QuantiNova RT buffer system | Read-through of difficult templates |
| RT Primer Mix | cDNA synthesis from all regions of transcripts, even from 5' regions |
See figures
Procedure
Genomic DNA removal and cDNA synthesis take only 20 minutes with the QuantiNova Reverse Transcription Kit. The kit includes everything you need for fast cDNA synthesis.
Purified RNA is briefly incubated in gDNA Removal Mix to effectively remove contaminating genomic DNA. In contrast to other methods, the RNA sample is then used directly for reverse transcription in combination with a master mix prepared from the Reverse Transcription Mix plus Reverse Transcription Enzyme. With the QuantiNova Reverse Transcriptase, RNA can be transcribed at low temperatures, even through complex secondary structure. This ensures that the RNA will stay intact as the reaction takes place at 42°C and is then inactivated at 85°C. Additional steps for RNA denaturation or RNase H digestion are not necessary.
The internal control (IC) RNA can be transcribed simultaneously with the RNA sample and is intended to report the presence of inhibitors, instrument or chemistry failures or errors in assay setup.
The QuantiNova IC Probe Assay (Max label, cat. no. 205813) or QuantiNova IC Probe Assay Red 650 (Cy5 analog label, cat. no. 205824) are used for probe based detection procedures of the Internal Control RNA or DNA provided in the QuantiNova Reverse Transcription Kit, the QuantiNova Probe RT-PCR Kit (one-step RT-PCR) or the QuantiNova Pathogen +IC Kit.
The QuantiNova IC SYBR Green Assay (cat. no. SBH1218551) is used in conjunction with the Internal Control RNA provided in the QuantiNova Reverse Transcription Kit (two-step RT-PCR), the QuantiNova SYBR Green RT-PCR Kit (one-step RT-PCR) and the QIAcuity OneStep Eva Green Kit. It permits assessment of reverse transcription/amplification performance using the QuantiNova SYBR Green PCR Kit (two-step RT-PCR), the QuantiNova SYBR Green RT-PCR Kit (one-step RT-PCR), the QIAcuity OneStep Eva Green Kit or the QuantiNova LNA PCR Custom Panels.
For a streamlined workflow, we recommend combining QuantiNova Kits with our predesigned QuantiNova real-time PCR assays or panels. These offer precise quantification of mRNA or long non-coding RNA transcripts, regardless of abundance. Choose from a wide range of predesigned human, mouse and rat primer sets, or customize your own assays and panels using our advanced design tools.
Our QuantiNova LNA PCR and QuantiNova LNA Probe PCR Assays utilize LNA technology for enhanced sensitivity, enabling unbiased gene expression profiles and faster scientific insights.
Note: The QuantiNova IC SYBR Green Assay was formerly available as QuantiTect Primer Assay for SYBR based detection of IC RNA (cat. no. QT02589307).
Applications
Supporting data and figures
Efficient removal of contaminating gDNA ensures precise quantification of transcripts.