qBiomarker Somatic Mutation PCR Arrays
For rapid and accurate profiling of the somatic DNA mutation status of gene panels
For rapid and accurate profiling of the somatic DNA mutation status of gene panels
Cat. No. / ID: 337021
qBiomarker Somatic Mutation PCR Arrays are translational research tools that allow rapid and accurate profiling of the somatic mutation status for important genes related to a biological pathway or disease. Mutations are selected from comprehensive somatic mutation databases (e.g., COSMIC) and peer-reviewed scientific literature based on their clinical or functional relevance and frequency of occurrence.
qBiomarker Somatic Mutation PCR Arrays show a high degree of sensitivity — even with formalin-fixed paraffin-embedded (FFPE) samples (see figures " High degree of sensitivity" and " Sensitivity of qBiomarker Somatic Mutation PCR Arrays with FFPE samples").
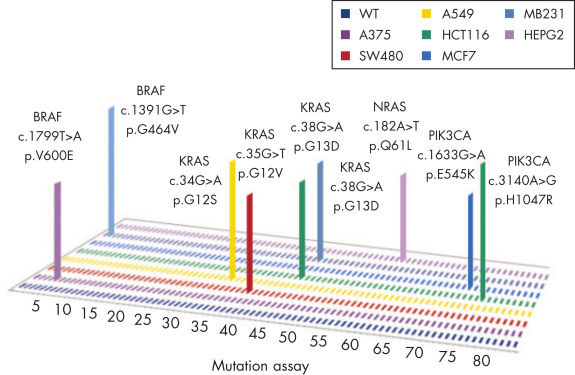
qBiomarker Somatic Mutation PCR Arrays have utility in the detection of mutations in cell lines or research samples, which is critical for toxicological, drug development, and cancer studies (see figure " Profiling of common cancer cell lines for somatic mutation status").
Simply mix the DNA template with the ready-to-use PCR mastermix, aliquot equal volumes to each well of the same plate, and then run the real-time PCR cycling program. qBiomarker Somatic Mutation PCR Arrays are compatible with all ABI, Bio-Rad, Eppendorf, Roche, and Stratagene instruments.
qBiomarker Somatic Mutation PCR Arrays are available in 96-well and 384-well plates and are used to detect mutations related to a disease state or pathway, plus gene copy number controls for normalization. Each qBiomarker Somatic Mutation PCR Array also includes control elements for general PCR performance.
Data can be analyzed using the available Excel-based data analysis templates. Data analysis is based on either the ∆∆CT or Average ∆CT method.
Arrays are available in a variety of formats, all with mastermix included:
qBiomarker Somatic Mutation PCR Arrays are highly suited for the rapid and accurate profiling of mutations for a pathway- or disease-focused set of genes and key downstream and associated signaling genes.