✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 51804
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Efficient lysis of bacteria and fungi from stool and gut samples
- Up to 20-fold more DNA compared to alternative methods
- Yield higher alpha diversity in sequencing compared to other methods
- Recovery of inhibitor-free DNA for direct use in next-generation sequencing applications
- Automate workflows on the QIAcube Connect
Product Details
Extracting microbial DNA from stool and gut samples can be challenging. Researchers will isolate higher yields of pure microbial DNA from stool and gut samples more efficiently with our new QIAamp PowerFecal Pro DNA Kit.
The QIAamp Power Fecal Pro DNA Kit builds and improves on our original PowerFecal technology utilizing a novel bead tube and optimized chemistry for more efficient lysis of bacteria and fungi. Inhibitor removal has been streamlined, allowing researchers to eliminate challenging inhibitors from stool and gut samples faster. This results in total DNA that reveals higher alpha diversity through observed operational taxonomic units (OTUs) in downstream testing.
Isolation of DNA using the QIAamp PowerFecal Pro DNA Kit can be automated on the QIAcube Connect.
Want to try this solution for the first time? Request a quote for a trial kit.
Performance
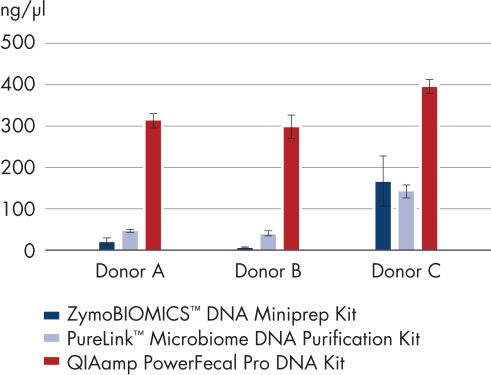
Using improved technologies, the new QIAamp PowerFecal Pro Kit allows researchers to obtain higher yields of DNA from stool and gut samples compared with alternative methods (see Higher yields of DNA with the QIAamp PowerFecal Pro DNA Kit).
Improved yields of higher quality DNA are also achieved when comparing the QIAamp PowerFecal Pro DNA Kit to the first generation QIAamp PowerFecal DNA Kit when running stool and fungal samples (see The new QIAamp PowerFecal Pro DNA Kit enables the isolation of larger amounts of high quality DNA).
See figures
Principle
The streamlined Inhibitor Removal Technology® (IRT) used by the new QIAamp PowerFecal Pro DNA Kit is highly efficient at removing inhibitors while still decreasing sample processing time. The QIAamp PowerFecal Pro DNA Kits also showed the highest A260/A280 ratios, near 1.8, of any other commercially available kit, indicating the absence of inhibitors. Less variability was also observed between the samples processed with QIAamp PowerFecal Pro DNA Kits (see Highly pure DNA isolated using the new QIAamp PowerFecal Pro DNA Kit). Researchers using the QIAamp PowerFecal Pro DNA Kit can identify a greater bacterial diversity with increased OTUs from stool samples compared with the first-generation QIAamp PowerFecal Kit and other commercially available kits (see Greater total number of bacterial OTUs isolated with the new QIAamp PowerFecal Pro DNA Kit).
See figures
Procedure
Samples processed with the QIAamp PowerFecal Pro DNA Kit are added to a bead beating tube. Rapid and thorough homogenization occurs using mechanical and chemical methods. Once cells are lysed, our patented IRT is used to remove inhibitors. Total genomic DNA is captured on a silica membrane in a spin-column format. DNA is then washed and eluted, ready for downstream applications.
Supporting data and figures
Higher yields of DNA with the QIAamp PowerFecal Pro DNA Kit.