Ni-NTA Fast Start Kit
E. coli溶解物からの組み換えHisタグ付きタンパク質の精製および検出用
E. coli溶解物からの組み換えHisタグ付きタンパク質の精製および検出用
✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
Cat. No. / ID: 30600
✓ オンライン注文による24時間年中無休の自動処理システム
✓ 知識豊富で専門的な製品&テクニカルサポート
✓ 迅速で信頼性の高い(再)注文
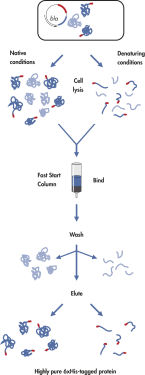
Ni-NTA Fast Start Kitは、6個以上のヒスチジン残基(連続または交互)のアフィニティタグ(Hisタグ)を含むタンパク質に対する特許取得済みのNi-NTA(ニッケル-ニトリロ三酢酸)樹脂の優れた選択性に基づいています。この技術により、天然または変性条件下で、ほぼすべてのHisタグ付きタンパク質をワンステップ精製できます。ニッケルイオンに対して4つのキレート部位を持つNTAは、金属イオンとの相互作用に利用できる部位が3つしかない金属キレート精製システムよりもしっかりとニッケルに結合します。余剰なキレート部位がニッケルイオンの浸出を防ぐため、他の金属キレート精製システムを使用した場合よりも高純度のタンパク質調製物が得られます。Hisタグは、キットに付属のPenta·His Antibodyによって認識されるエピトープも生成します。
Ni-NTA Fast Start Kitは、以下を含むあらゆるアプリケーションに適した6xHisタグ付きタンパク質を高い信頼性でワンステップ精製します。
| Features | Specifications |
|---|---|
| Applications | プロテオミクス |
| Support/matrix | Ni-NTAマトリックス |
| Processing | 手動 |
| Binding capacity | 5~20 mg/ml |
| 特殊機能 | Ni-NTA技術 |
| Gravity flow or spin column | 自然落下 |
| Start material | 細胞溶解物 |
| Tag | 6xHisタグ |
| Yield | カラムあたり<10 mgタンパク質 |