✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 78404
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Efficient isolation of high-quality microbial RNA
- Easy to use protocol for isolating total RNA from difficult microbiome samples
- Highly pure RNA isolation with Inhibitor Removal Technology
- Automatable on QIAcube Connect
Product Details
With the RNeasy Powerfecal Pro Kit, you can isolate total RNA from stool, sludge and wastewater samples high in PCR inhibitors. The kit uses QIAGEN’s second-generation Inhibitor Removal Technology® (IRT), resulting in high yields of highly pure RNA, which can be used immediately in downstream applications, including RT-PCR, qPCR and next-generation sequencing (e.g., RNA-seq or metatranscriptome). Proven bead beating technology and lysis chemistry complete the easy to use, proprietary protocol.
Isolation of RNA using the RNeasy PowerFecal Pro Kit can be automated on the QIAcube Connect.
Want to try this solution for the first time? Request a quote for a trial kit .
Performance
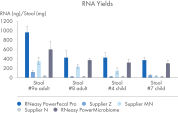
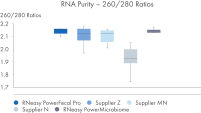
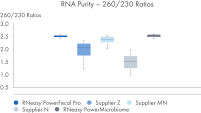
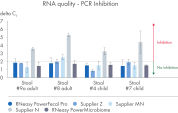
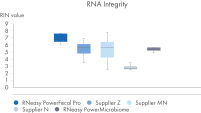
RNeasy PowerFecal Pro kit enables high yield and high-purity RNA from diverse microbiome samples (see figures RNA Yields – QIAexpert Quantitifcation, RNA Purity 260/280 Ratios, RNA Purity 260/230 Ratios). The kit allows for efficient removal of PCR inhibitors for pure RNA with low inhibitor co-isolation (see figure RNA Quality – PCR Inhibition) and high RNA quality (See figures RIN Value, 23S/16S Ratio).
See figures
Principle
The RNeasy PowerFecal Pro Kit efficiently isolates total RNA from diverse starting material such as stool, gut, sludge or wastewater. The RNeasy PowerFecal Pro Kit provide the highest-quality RNA with minimum copurification of DNA. The kit can be used for automated RNA isolation on QIAcube Connect.
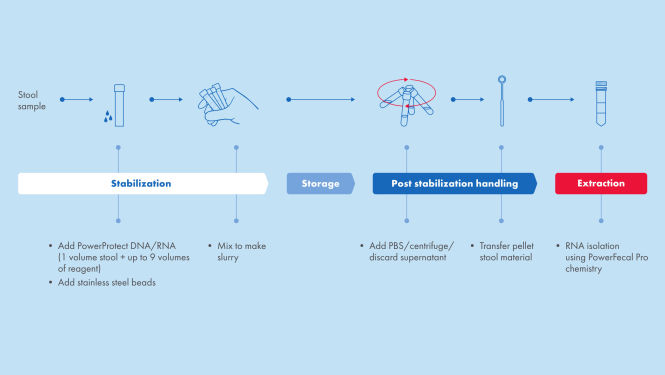
It is important to preserve your stool samples shortly after collection, to maintain the integrity of your RNA. With PowerProtect DNA/RNA for stabilization, we provide a comprehensive solution, from collection to extraction (see figure Sample stabilization and RNA extraction).
See figures
Procedure
We recommend to start with 50–200 mg of stool. Homogenize each sample in a 2 ml bead beating tube containing a mixture of lysis beads. The mechanical collisions between beads combined with the chemical disruption of cell membranes allow for lysis of host and microbial cells. IRT is then used to remove substances that are commonly found in stool samples, which inhibit PCR and other downstream applications. Then pass the lysate through a MB RNA Spin Column. Remove the DNA using on-column DNase digestion. Then use a wash solution to remove the enzyme and any digested nucleic acids (see figure RNeasy PowerFecal Pro Procedure). Elute the RNA with RNase-free water. It is now ready to use in any downstream applications.
Sample storage and preservation
The yield and integrity of nucleic acids isolated from microbes in stool is greatly influenced by the state of the digestive system, diet of the individual, and the length of time between collection of the sample and preservation. To optimize the quality of nucleic acids from stool, process the sample as quickly as possible after collection. The PowerProtect DNA/RNA reagent stabilizes stool samples at room temperature.
Automated processing on the QIAcube Connect
RNA purification with the RNeasy PowerFecal Pro kit can be fully automated on QIAcube Connect or the classic QIAcube.
The QIAcube Connect instrument is designed for low-throughput automation of the sample preparation workflow based on spin column technology. The automated protocol follows the same steps as a manual procedure (i.e., lyse, bind, wash, and elute) processing up to 12 samples per run.
QIAcube Connect is designed to perform automated purification of nucleic acids and proteins in molecular biology applications. The system is intended for use by professional users trained in molecular biological techniques and the operation of QIAcube Connect.
See figures
Applications
The RNeasy PowerFecal Pro Kit enables isolation of high-quality RNA that can be used immediately in downstream applications, including RT-PCR, qPCR, digital PCR and next-generation sequencing (e.g., RNA-seq or metatranscriptome).
Supporting data and figures
Sample stabilization and RNA extraction
Sample stabilization with PowerProtect DNA/RNA Kit and RNA isolation with RNeasy PowerFecal Pro Kit
Specifications
| Features | Specifications |
|---|---|
| Sample types | Stool, gut material, sludge, wastewater |
| Format | 2 ml Tubes |
| Sample size | 50-200 mg |
| Throughput | 1-50 samples |
| Processing | Bead beating tube 2 ml |
| Storage temperature | Store CD2 and lyophilized DNase I at 2-8°C All other kit reagents and components should be stored at room temperature (15-25°C) |