Cat. No. / ID: 80254
Features
- Improved, second generation Inhibitor Removal Technology (IRT)
- Separates RNA and DNA from the same sample into different eluates in one procedure
- Optimized lysis for increased DNA and RNA yields
- RNA and DNA for metagenomic and metatranscriptomic analysis
Product Details
With the AllPrep PowerFecal Pro DNA/RNA Kit you can extract both DNA and RNA from the same stoll or gut sample using a proprietary protocol with improved Inhibitor Removal Technology (IRT). Second generation IRT is effective with samples containing inhibitory substances, such as polysaccharides, heme compounds, and bile salts, which are commonly found in stool. An optimized lysis chemistry combined with bead beating technology allow for high yields of high-quality DNA and RNA that can be used immediately in downstream applications, including PCR, RT-PCR, qPCR, and next-generation sequencing (NGS) (16S, metagenome, and metatranscriptome).
If you want to learn more about our complete stabilization and extraction solution, visit the PowerProtect DNA/RNA Kit webpage.
Want to try AllPrep PowerFecal Pro DNA/RNA Kit for the first time? Request a quote for a trial kit .
Performance
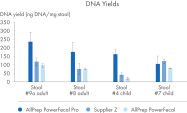
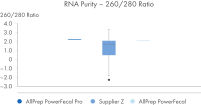
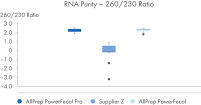
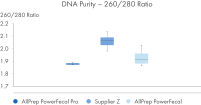
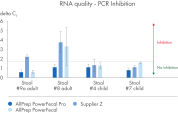
AllPrep PowerFecal Pro DNA/RNA kit enables high yield of high- purity nucleic acids simultaneously extracted from the same stool sample (see figures RNA Yields – QIAexpert Quantitifcation, DNA Yields -QIAexpert Quantification, RNA Quality 260/280 Ratio, RNA Quality 260/230 Ratio, DNA Quality 260/280 Ratio, DNA Quality 260/230 Ratio). The kit efficiently purifies DNA and RNA from stool samples that are inherently rich in PCR inhibitors due to good inhibitor depletion (see figure RNA Quality – PCR Inhibition). The kit allows for high yields of high-quality RNA (See figures RIN Value, 23S/16S Ratio). The DNA and RNA are suitable for NGS analysis such as microbial operational taxonomic unit (OTU) analysis or comparative metagenomic and metatranscriptomic studies (see figure Operational Taxonomic Units). Comprehensive microbial abundance is demonstrated (see figures Alpha Diversity and Beta Diversity).
See figures
Principle
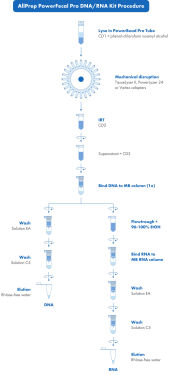
The AllPrep PowerFecal Pro DNA/RNA Kit is designed to purify microbial DNA and RNA simultaneously from stool samples. The nucleic acids are obtained from the entire sample during the same process (see figure All Prep PowerFecal Pro DNA/RNA Procedure). The AllPrep PowerFecal Pro DNA/RNA Kit integrates three QIAGEN technologies for effective DNA and RNA extraction: IRT in the initial lysis, selective binding of double-stranded DNA and RNeasy® technology for RNA isolation with minimum copurification of DNA.
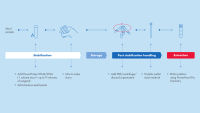
To protect the integrity of the nucleic acids, we recommend you preserve your stool sample as soon as possible after collection. With PowerProtect DNA/RNA for storage and stabilization, we provide a comprehensive solution (see figure Sample stabilization and extraction of DNA/RNA), from collection to extraction.
See figures
Procedure
We recommend you start with 50–200 mg of stool. Homogenize each sample in a 2 ml bead beating tube containing a mixture of lysis beads. The combination of mechanical bead beating and proven lysis chemistry facilitates optimized lysis of host and microbial cells. Remove common inhibitors found in stool samples, and which interfere with PCR and other downstream applications with second generation IRT. Pass the lysate through an MB Spin Column in combination with a high-salt buffer, to allow selective, efficient genomic DNA binding. Wash and elute the DNA. It is ready for PCR analysis and other downstream applications, including qPCR and NGS.
Add Ethanol to the flow-through from the MB Spin Column to create appropriate RNA-binding conditions. Transfer the sample to an MB RNA Spin Column, for total RNA binding to the membrane. Wash and elute the RNA in RNase-free water. It is now ready for downstream applications including RT-PCR, qPCR and NGS.
Sample storage, stabilisation and preservation
The time between sample collection and preservation is among the main conditions to influence the yield and integrity of nucleic acids isolated from microbes in stool samples, together with the state of the digestive system and diet of the individua. We recommend that you process the sample as quickly as possible after collection, to optimize the quality of nucleic acids from stool. The PowerProtect DNA/RNA reagent enables stabilizes of stool samples at room temperature.
Applications
The AllPrep PowerFecal Pro DNA/RNA Kit enables simultaneous extraction of high-quality DNA and RNA. The DNA can be used immediately in PCR analysis and other downstream applications, including qPCR and NGS. The RNA can be used in downstream applications including RT-PCR, qPCR, digital PCR and next-generation sequencing (e.g., RNA-seq or metatranscriptome).
Supporting data and figures
AllPrep PowerFecal Pro DNA/RNA Procedure
DNA and RNA simultaneous extraction workflow with the AllPrep PowerFecal Pro DNA/RNA Kit
Specifications
| Features | Specifications |
|---|---|
| Sample types | Stool, gut material |
| Format | 2 ml tubes |
| Sample size | 50-200 mg |
| Processing | PowerBead Pro Tubes (mix of zirconia beads) |
| Storage temperature | Store CD2 at 2-8°C. All other kit reagents and components should be stored at room temperature (15-25°C) |