✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 59695
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Convenient, ready-to-use, quality-controlled DNA
- Bisulfite-converted DNA for control experiments
- Suitable for all methylation analyses
Product Details
Performance
See figures
Principle
See figures
Procedure
Standardized workflows in epigenetics
Accessing epigenetic information is of prime importance for many areas of biological and medical research — particularly oncology, but also stem cell research and developmental biology. However, the analysis of changes in DNA methylation is challenging, due to the lack of standardized methods for providing reproducible data particularly from limited sample material. With its newly introduced EpiTect solutions, QIAGEN makes available standardized, pre-analytical and analytical solutions from DNA sample collection, stabilization and purification, to bisulfite conversion and real-time or end-point PCR methylation analysis or sequencing (see figure " Standardized workflows in epigenetics").
See figures
Applications
EpiTect Control DNAs are suitable for use in control reactions for all types of methylation analyses, including:
- Evaluating primer specificity for MSP
- Quantification standard for HRM and MethyLight PCR
- Evaluating primer and probe specificity for MethyLight PCR
- Checking efficiency of bisulfite conversion reactions
Supporting data and figures
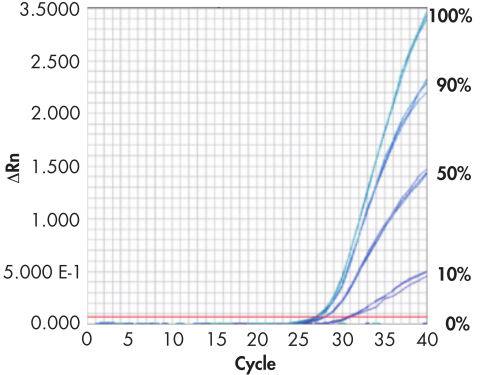
Real-time PCR standards.
EpiTect Methylated Control DNA and EpiTect Unmethylated Control DNA (both pre-bisulfite converted) were mixed to give 100%, 90%, 50%, 10%, and 0% methylated DNA. EpiTect MethyLight Assays for the human PITX2 gene were run in triplicate, using 10 ng of each DNA sample. The results show that EpiTect Control DNA can be used as a methylation standard for the quantification of unknown DNA samples.
Specifications
| Features | Specifications |
|---|---|
| Applications | End point Methylation Specific PCR (MSP), real-time methylation specific PCR |
| Number of reactions | for 1000 control PCRs |
| Concentration | 10 ng/µl (1 µg in total) excepted the unmethylated control DNA: 50 ng/µl (10 µg in total) |