AllStars Negative Control siRNA
For negative control RNAi experiments
For negative control RNAi experiments
The performance of AllStars Negative Control siRNA was validated by experiments shown in the table.
| Test type | Test name | Purpose | Result for AllStars Negative Control siRNA |
|---|---|---|---|
| Genomewide analysis | Affymetrix GeneChip arrays | Nonspecific regulation of gene expression | Minimal number of genes regulated |
| Cell-based assay | Live-cell nuclei staining | Nuclear size | Normal |
| Cell-based assay | Cell number | Proliferation rates | Unchanged |
| Cell-based assay | Nucleotide incorporation | DNA synthesis rates | Unchanged |
| Cell-based assay | Live-cell dye exclusion | Cytotoxic effects | Unchanged |
| Cell-based assay | DNA staining | Cell-cycle distribution | Normal |
| RISC-incorporation analysis (HeLa and MCF-7 cells) | Reporter construct transfection | Determine whether siRNA is incorporated into RISC (a valid negative control should enter RISC) | Incorporated into RISC |
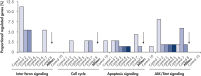
Genomewide analysis was performed to test the level of nonspecific effects on gene expression after transfection of multiple negative control siRNAs. Multiple negative control siRNAs from different origins were transfected into MCF-7, K562, and primary HUVEC cells using HiPerFect Transfection Reagent. These included nonsilencing siRNAs (with no homology to mammalian genes), scrambled siRNAs (siRNAs with the same base composition as the gene-specific siRNA but with an altered sequence), and siRNAs targeting artificial reporter genes. Subsequently, expression profiling of the whole human genome was performed with Affymetrix GeneChip arrays. AllStars Negative Control siRNA consistently resulted in the lowest number of nonspecifically regulated genes, making it a highly suitable negative control. In contrast, other negative control siRNAs resulted in nonspecific regulation of many genes from important cellular pathways (see figure " Low nonspecific effects on expression").
Live-cell nuclei staining was used to measure nuclear size. Changes in size could be an indication of cell-cycle disturbance or growth inhibition. A range of negative control siRNAs of different types were tested. AllStars Negative Control siRNA provided the best results of all the controls tested. Transfection of AllStars Negative Control siRNA did not result in any change in nuclear size compared with untransfected cells. In contrast, transfection of another negative control siRNA (Control 1) resulted in enlarged nuclei (see figure " Nuclear size phenotype unaffected").
Cell number was assessed after transfection of a range of negative control siRNAs to determine whether cells were proliferating normally. Almost no difference in cell number was observed between untransfected cells and cells transfected with AllStars Negative Control siRNA. In contrast, cell number was significantly reduced after transfection with other negative control siRNAs tested, such as Control 1, indicating that these siRNAs resulted in a growth defect phenotype (see figure " Cell number unaffected").
Nucleotide incorporation was measured to determine DNA synthesis rates in untransfected HCT-116 cells and in HCT-116 cells transfected with a range of different negative control siRNAs. Nucleotide incorporation was measured by examining the uptake of Bromodeoxyuridine (BrdU), a base analog of thymidine that substitutes for thymidine during DNA replication and is incorporated into newly synthesized DNA. Changes in DNA synthesis rates could indicate altered cell growth or cell cycle. Cells transfected with AllStars Negative Control siRNA showed a BrdU-incorporation rate that was very similar to that of untransfected cells. However, another negative control siRNA tested (Control 1) resulted in an altered profile with a lower level of DNA synthesis, which indicates that this siRNA affects cell growth or the cell cycle (see figure " Normal DNA synthesis phenotype").
Live-cell dye exclusion was used to measure potential cytotoxic effects of a range of negative control siRNAs. Results showed that cells transfected with AllStars Negative Control siRNA and untransfected cells had a similar number of living and dead cells. In contrast, other negative control siRNAs resulted in an increase in cytotoxicity (see figure " No increase in cytotoxic effects").
DNA staining after cell fixation was used to measure cell-cycle distribution (the amount of cells in G1/G0, S, and G2 phases of the cell cycle). After transfection of AllStars Negative Control siRNA, the proportion of cells in each phase was similar to that observed for untransfected cells (see figure " Normal cell-cycle distribution"). This result demonstrates that AllStars Negative Control siRNA does not adversely affect the cell cycle.
For accurate negative control RNAi experiments, the negative control siRNA should be incorporated into RISC (RNA-Induced Silencing Complex). This means that the control siRNA goes through the same biological process as the gene-specific siRNA and enables comparison of data from gene-specific siRNA with data from negative control siRNA to confidently determine results that are due to target gene knockdown.
Experiments were performed as follows.
Cotransfection of the reporter construct with a noncomplementary siRNA resulted in strong expression of the fluorescent protein and the His tag. When the construct was cotransfected with AllStars Negative Control siRNA, the siRNA knocked down expression from its complementary sequence, resulting in degradation of the entire mRNA transcript encoding the fluorescent reporter gene, the His tag, and the siRNA target sequence. The mRNA degradation resulted in knockdown of the fusion protein (see figures " AllStars Negative Control siRNA is incorporated into RISC" and " Western analysis shows AllStars Negative Control siRNA enters RISC"). To achieve the knockdown observed, AllStars Negative Control siRNA must have entered RISC.
 Nuclear size phenotype unaffected.
Nuclear size phenotype unaffected.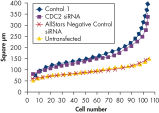 Cell number unaffected.
Cell number unaffected.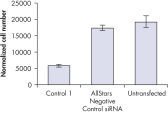 Normal DNA synthesis phenotype.
Normal DNA synthesis phenotype. No increase in cytotoxic effects.
No increase in cytotoxic effects. Normal cell-cycle distribution.
Normal cell-cycle distribution. Reporter construct for RISC-incorporation experiment
Reporter construct for RISC-incorporation experiment AllStars Negative Control siRNA is incorporated into RISC.
AllStars Negative Control siRNA is incorporated into RISC.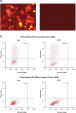 Western analysis shows AllStars Negative Control siRNA enters RISC.
Western analysis shows AllStars Negative Control siRNA enters RISC.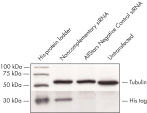
Transfection of a negative control siRNA is essential in every RNAi experiment. Results from the negative control should be compared with results from untransfected cells. Gene expression and phenotype should ideally be similar in both untransfected cells and cells transfected with negative control siRNA. If altered expression or phenotype are observed in cells transfected with negative control siRNA, these changes are nonspecific – they are due to transfection procedures or siRNA toxicity and not sequence complementarity. Nonspecific effects should be minimal to ensure reliable RNAi/miRNA results.
Results from the negative control can also be compared with results from the gene-specific siRNA/miRNA under study. This comparison allows the researcher to pinpoint the effects of target-gene knockdown on gene expression and phenotype, because the negative control sample has undergone the same biological process, with the only difference being the siRNA/miRNA sequence.
Results from AllStars Negative Control siRNA can be used as follows:
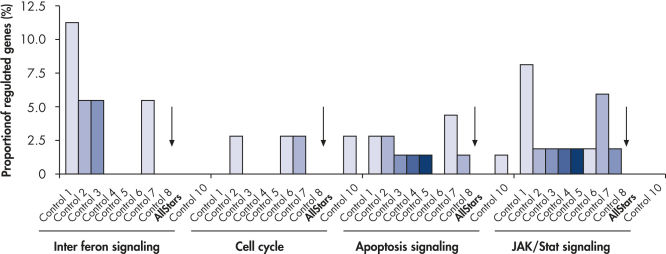
Multiple negative control siRNAs (Control 1– Control 10) were transfected in triplicate into MCF-7 cells. After incubation, cRNA was prepared and hybridized to Affymetrix human U133 GeneChip arrays. Regulated genes were identified as genes that showed at least a 1.5-fold change in expression (both upregulated and downregulated) compared to untransfected cells. Ingenuity pathway analysis software was used to determine the proportion of regulated genes in each pathway compared to the total number of genes identified as central to that pathway. Where a bar appears in the figure, this means that genes in the pathway were regulated by the siRNA. If every pathway gene was regulated, the relative proportion would be 100%. Lower bars therefore indicate a lower relative proportion of regulated genes within that pathway. Where no bar appears, no genes of the pathway were regulated by the siRNA. AllStars Negative Control siRNA (indicated with arrow) resulted in the lowest number of regulated genes. In contrast, other control siRNAs resulted in higher numbers of regulated genes from important cellular pathways.
| Features | Specifications |
|---|---|
| Design | Predesigned/validated by Affymetrix GeneChip Array and cell-based assays |
| Species | Human, mouse, rat |
| Scale or yield | 5 nmol, 20 nmol |
| Format | Tube |
| Target sequence provided | No |
| Modification | Yes |