✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Cat. No. / ID: 31314
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- 每个离心柱可在短短 15 分钟内纯化多达 300 μg His 标签蛋白
- 在天然和变性条件下进行纯化
- 一步实现高达 95% 的均一性
- 用于快速自动化或手动处理的即用型离心柱
Product Details
Ni-NTA 硅胶将 Ni-NTA 与大孔硅胶载体材料相结合,并经过优化以抑制非特异性疏水相互作用。Ni-NTA Spin Kit 内附带以及可单独供应的 Ni-NTA Spin Column(His 蛋白纯化离心柱)提供了便利的微离心形式的 Ni-NTA 硅胶,能够很容易地同时制备多个样本。它们为工程化蛋白的功能筛选、表达全长翻译产物的克隆选择以及表达水平的比较提供了一种简单的方法。每个离心柱可以纯化多达 300 µg His 标签蛋白。与所有 Ni-NTA 基质一样,Ni-NTA 离心柱可用于在天然或变性条件下的一步法蛋白质纯化。Ni-NTA Spin Kit 是用于 His 标签蛋白离心纯化的完整试剂盒。它可以在 QIAcube Connect 上实现自动化(参见图片“ QIAcube Connect”)。
See figures
Performance
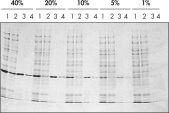
Ni-NTA Spin Column(His 蛋白纯化离心柱)也包含在 Ni-NTA Spin Kit 中,允许在不同表达水平下(参见图片 “不同表达水平下的纯化”)进行可重复的快速自动纯化(参见图片 “可重复的自动纯化”)。
See figures
Principle
QIAexpress Ni-NTA Protein Purification System,包括 Ni-NTA Spin Columns 和 Ni-NTA Spin Kit,基于专利 Ni-NTA(镍-次氮基三乙酸)树脂,它对含有六个或更多组氨酸残基(即 His 标签)的亲和标签的蛋白质具有显著的选择性。该技术允许在天然或变性条件下从任何表达系统中一步纯化几乎任何 His 标签蛋白。NTA 具有四个镍离子螯合位点,与只有三个可用于与金属离子相互作用位点的金属螯合纯化系统相比,其与镍的结合更加紧密。这个额外的螯合位点能够防止镍离子浸出,与其他金属螯合纯化系统相比,能够产生更强的结合能力并得到更高纯度的蛋白质制剂。QIAexpress 系统可用于从包括杆状病毒、哺乳动物细胞、酵母和细菌在内的任何表达系统中纯化 His 标签蛋白。
Procedure
His 标签蛋白的纯化包括 4 个步骤:细胞裂解、结合、洗涤和洗脱(参见图片“使用 Ni-NTA 蛋白纯化系统进行 Ni-NTA 离心柱纯化”)。使用 QIAexpress 系统进行重组蛋白纯化不依赖于蛋白质或 His 标签的三维结构。这允许在天然或变性条件下从稀释溶液和粗制裂解物中一步纯化蛋白质。将最多 600 μl 细胞裂解物装载至 Ni-NTA 离心柱。快速离心 2 分钟即可使标签蛋白结合至 Ni-NTA 硅胶,而大多数非标签蛋白都会流过硅胶。经过一次洗涤步骤后,在温和条件下(例如 pH 降低至 5.9,或加入 100-500 mM 咪唑)以 100-300 μl 的体积洗脱纯化的蛋白质。通常不需要去除 His 标签,因为它很小,而且很少具有免疫原性。纯化后的蛋白质可以立即使用。可在大约 30 分钟(手动程序)或大约 60 分钟(QIAcube Connect 自动化程序)内从多种小规模表达培养物中纯化出蛋白质。可使用强变性剂和洗涤剂进行受体、膜蛋白和形成包涵体的蛋白质的高效溶解和纯化。洗涤缓冲液中可以包括能够高效去除非特异性结合污染物的试剂(参见表格)。通过加入 100-250 mM 咪唑作为竞争物或通过降低 pH 在温和条件下洗脱纯化的蛋白质。
与 Ni-NTA–His 相互作用兼容的试剂:
- 6 M 盐酸胍
- 8 M 尿素
- 2% Triton X-100
- 2% Tween 20
- 1% CHAPS
- 20 mM β-ME
- 10 mM DTT
- 50% 甘油
- 20% 乙醇
- 2 M NaCl
- 4 M MgCl2
- 5 mM CaCl2
- ≤20 mM 咪唑
- 20 mM TCEP
See figures
Applications
QIAexpress Ni-NTA Protein Purification System,包括 Ni-NTA Spin Columns 和 Ni-NTA Spin Kit,提供了适用于任何应用的可靠一步法蛋白质纯化,包括:
- 结构和功能研究
- 结晶用于三维结构测定
- 涉及蛋白质–蛋白质和蛋白质–DNA 相互作用的检测
- 免疫接种产生抗体
| 特点 | Ni-NTA Spin Columns | Ni-NTA Spin Kit |
| 应用 | 蛋白质组学 | 蛋白质组学 |
| 微珠大小 | 16–24 µm | 16–24 µm |
| 结合能力 | 每个离心柱最多 300 µg | 每个离心柱最多 300 µg |
| 重力流或离心柱 | 离心柱 | 离心柱 |
| 处理 | 自动化/手动 | 自动化 |
| 规模 | 小规模 | 小规模 |
| 特殊特点 | 低通量筛选 | 一步实现高达 95% 的均一性 |
| 起始材料 | 细胞裂解物 | 细胞裂解物 |
| 载体/基质 | 大孔硅胶 | 大孔硅胶 |
| 标签 | 6xHis 标签 | 6xHis 标签 |
Supporting data and figures
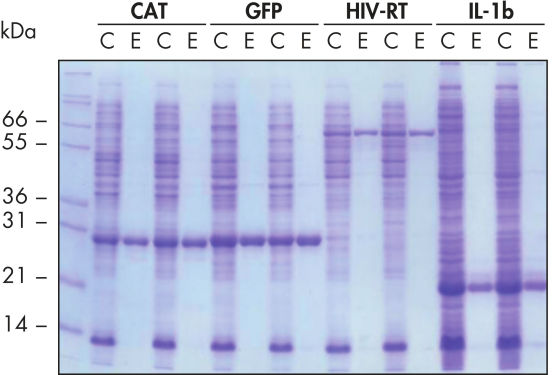
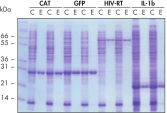
可重复的自动纯化。
在天然条件下,使用 Ni-NTA Spin Columns 从源自 5 ml LB 培养物的澄清 E. coli 细胞裂解物中一式两份纯化所示的蛋白质(手动操作或在 QIAcube 上使用自动化程序)。CAT:氯霉素乙酰转移酶;GFP:绿色荧光蛋白;HIV-RT:人类免疫缺陷病毒逆转录酶;IL-1b:白细胞介素-1β。M:分子量标准;C:澄清的裂解物(每道加载 2 μl);E:洗脱馏分(每道加载 3 μl)。