✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
QIAseq Universal Normalizer Kit (24)
Cat. No. / ID: 180613
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Produces normalized, ready-to-sequence libraries at 4 nmol/L
- Streamlined 30-minute benchtop protocol
- Seamless integration into many NGS workflows including QIAseq as well as virtually all third-party library preparation workflows for Illumina
- Produces dsDNA for stable, longer term library storage
- Fume hood not required
Product Details
The QIAseq Normalizer Kits offer a fast and easy workflow to adjust many different types of libraries to a fixed concentration (4 nmol/L). Once normalized, libraries can be pooled at equal volumes to achieve homogenous read representation without the need for library quantification prior to sequencing. The QIAseq Normalizer Kits enable streamlined NGS workflows that are cost effective and have a shorter processing time compared to conventional methods using classical library quantification and pooling techniques.
The QIAseq Normalizer workflows are compatible with virtually all libraries that can be sequenced on Illumina sequencing instruments. This includes QIAGEN as well as third-party library preparation workflows for Illumina sequencing instruments.
Want to try the QIAseq Library Normalizer Kit for the first time? Request a quote for a trial kit.
Want to try the QIAseq Universal Normalizer Kit for the first time? Request a quote for a trial kit.
Performance
- Reliable normalization over a broad range of library concentrations from 15 nmol/L up to more than 300 nmol/L
- Low library concentration requirements: 15 nmol/L or more in 15 µl volume
- Full compatibility with various types of libraries: whole genome, whole transcriptome, target-enriched libraries
- Reproducible, normalized dsDNA libraries at 4 nmol/L
The QIAseq Normalizer Kits enable the user to:
- Reduce costs and maximize read utilization by homogeneous library representation
- Streamline the library to sequencing process by replacing time-consuming library quantification methods with the QIAseq Normalizer workflow
- Remove tedious PCR methods from their workflows
- Achieve qPCR-level accuracy library normalization in minimal time
- Store normalized dsDNA libraries for a longer term
Principle
QIAseq Normalizer Kits make use of modified library amplification primers and a chemistry based on magnetic beads to efficiently bind and release a defined amount of library molecules in a tightly controlled fashion. With a target concentration of approximately 4 nmol/L normalized libraries are tightly adjusted for balanced library pooling and optimal clustering on Illumina flow cells. The dsDNA library molecules are eluted from the beads at non-denaturing conditions and can be safely stored at -20°C for longer-term storage.
Cost-efficient multiplexed sequencing requires accurate pooling of libraries to achieve comparable sequencing depths for all libraries. Failure to combine libraries at equal ratios will lead to under- or overrepresentation of libraries in terms of sequencing yield. Underrepresented libraries will result in too few sequence reads, and overrepresented libraries will waste sequencing capacity. Today’s gold standard to enable library pooling at acceptable accuracies is to quantify each library prior to pooling.
Typical methods for library quantification include PCR (qPCR/dPCR), capillary gel electrophoresis and colorimetric DNA assays. While qPCR has proven to quantify libraries with high accuracy, it is laborious, time-intensive and costly to perform. In addition, pre-diluting libraries for qPCR and manually diluting libraries based on the assayed concentration provide more opportunity for error.
Procedure
The QIAseq Normalizer workflow follows a simple bind – wash – elute routine, which is identical for all compatible types of libraries. Normalized libraries are dsDNA and have a common concentration of approximately 4 nmol/L, which allows them to be directly used for pooling and sequencing.
Prior to normalization, libraries must be modified using the Normalizer Primer Mix. During a 2-cycle PCR in the presence of the Normalizer Primer Mix, modifications can be added to virtually any type of Illumina library with intact P5/P7 ends. P5/P7 ends are present on any finished library that can be sequenced on Illumina instruments.
Library preparation workflows that involve the ligation of full-length indexed-adapters enable in-process library modification and do not require an additional PCR step. Full-length adapters, such as QIAseq CDI/UDI Y-Adapters, contain full-length P5/P7 sequences. Therefore, libraries can be modified using the Normalizer Primer Mix during the library amplification step. The QIAseq Normalizer workflow is not compatible with PCR-free library preparation.
Applications
Libraries normalized with the QIAseq Normalizer Kits are directly ready for sequencing on an Illumina instrument.
The QIAseq Normalizer Kits are compatible with all QIAseq library preparation kits and virtually all third-party library preparation kits, as long as they are not intended for downstream hybrid capture.
Supporting data and figures
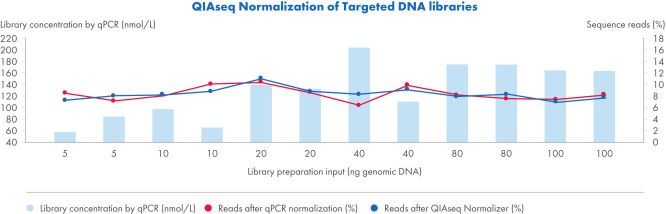
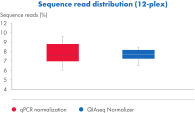
Achieve qPCR-level accuracy library normalization with the QIAseq Normalizer.
QIAseq Targeted DNA Panels library preparation from a broad range of gDNA input results in homogeneous read distribution after QIAseq normalization. The accuracy of library balancing is equal or superior to the performance achieved by qPCR.
Using 5–100 ng of gDNA, 12 QIAseq Targeted DNA libraries were prepared. All libraries were subjected to qPCR and diluted to 4 nmol/L before sequencing on the Ilumina MiSeq (V2 reagents, 2x150 bp paired-end). For comparison, the same libraries were normalized using the QIAseq Universal Normalizer Kit and sequenced on the Illumina MiSeq.