✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
REPLI-g WTA Single Cell Kit (24)
Cat no. / ID. 150063
✓ 24/7 automatic processing of online orders
✓ Knowledgeable and professional Product & Technical Support
✓ Fast and reliable (re)-ordering
Features
- Complete transcriptome coverage from just single cells
- Uniform WTA with negligible sequence bias due to MDA technology
- Optimized for use with new technologies, including NGS
- Amplification of total RNA or mRNA-enriched (poly A+) RNA
- Novel tool for cancer and stem cell research
Product Details
Performance
Complete transcriptome coverage, with low experimental variability
The REPLI-g WTA Single Cell Kit contains novel REPLI-g SensiPhi DNA Polymerase, as well as an optimized set of buffers and reagents for whole transcriptome amplification (WTA) from just single cells, up to 1000 cells, or equivalently small samples. Following efficient cell lysis, complete removal of genomic DNA (gDNA), and sensitive reverse transcription, the kit utilizes Multiple Displacement Amplification (MDA) to uniformly amplify cDNA across the entire transcriptome with negligible sequence bias (see figure Multiple Displacement Amplification (MDA) technology). cDNA amplified using the REPLI-g WTA Single Cell Kit demonstrates a high degree of reproducibility from cell to cell and experiment to experiment, in both next-generation sequencing (NGS) and in real-time PCR analysis of specific transcripts (see figure High level of experimental reproducibility). The amplified cDNA can be easily used in a variety of downstream applications (see figure REPLI-g amplified cDNA performs like gDNA in downstream experiments).Significant number of reads map to protein-coding RNA
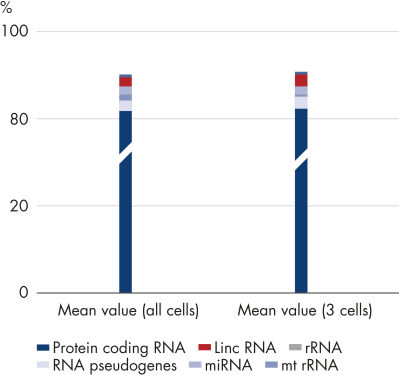
The ability to amplify mRNA-enriched RNA (poly A+) makes the REPLI-g WTA Single Cell Kit particularly suited for use in NGS to investigate effects on transcription regulation at the single-cell transcriptome level. Amplification of ribosomal RNA (rRNA), which generates more than 90% of NGS reads, is virtually eliminated, allowing generation of meaningful mRNA-Seq data. Following WTA using an mRNA-enrichment protocol, more than 80% of reads map to protein coding RNA (see figure High number of mappable reads from just 3 cells).Reliable detection of low-abundance transcripts
Single cell analysis can be challenging when transcript abundance varies greatly within a cell. For accurate results, it is essential that whole transcriptome amplification reliably amplifies all transcripts, regardless of their levels within the cell. The REPLI-g WTA Single Cell Kit is highly suitable for the analysis of even low-abundance transcripts from just single cells. Real-time PCR analysis of the REPLI-g amplified transcript abl-1 demonstrates that even low-copy transcripts can be reliably detected following amplification using the kit (see figure Highly reliable detection of even low-abundance transcripts from single cells). Unlike kits from other suppliers, the REPLI-g WTA Single Cell WTA Kit allows sensitive detection of all transcripts, from high- to low-abundance (see figure More sensitive detection of even low-abundance transcripts).For use in a wide variety of applications and research areas
The REPLI-g WTA Single Cell Kit efficiently generates and amplifies cDNA from single cells, such as tumor cells, stem cells, or sorted cells and from purified total or poly A+ RNA (10 pg – 100 ng), making the kit highly suited for a wide variety of research areas (see table).| Sample material (cells/total RNA) | Research area |
|---|---|
| Human/animal | Biomarker research (expression) |
| Stem cell research | |
| Analysis of circulating fetal cells | |
| Mosaicism studies | |
| Genetic predisposition studies | |
| Typing of transgenic animals | |
| Cancer | Somatic genetic variant analysis |
| Tumor progression | |
| Tumor stem cells/evoluation | |
| Analysis of circulating tumor cells |
Principle
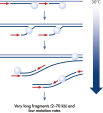
The REPLI-g WTA Single Cell Kit has been specifically designed to reliably investigate effects on transcription regulation at the single-cell transcriptome level. It provides highly uniform amplification across the entire transcriptome, with negligible sequence bias. Whole transcriptome amplification from single cells that is provided by the REPLI‑g WTA Single Cell Kit complements the respective whole genome amplification kit (REPLI-g Single Cell Kit). The method is based on MDA technology, which carries out isothermal cDNA amplification utilizing a uniquely processive DNA polymerase capable of replicating up to 70 kb without dissociating from the cDNA template (see figure Multiple Displacement Amplification (MDA) technology).
Unique components of the REPLI-g WTA Single Cell Kit
- All of the kit’s enzymes and amplification components undergo a unique, controlled decontamination procedure to ensure elimination of REPLI‑g amplifiable contaminating DNA or RNA. Following this process, the kits undergo stringent quality control to ensure complete functionality.
- The innovative lysis buffer effectively stabilizes cellular RNA. This ensures that the resulting RNA accurately reflects the in vivo gene expression profile.
- All enzymatic steps have been specifically developed to enable efficient processing of RNA for accurate amplification. For example, these processes include effective gDNA removal prior to cDNA synthesis.
- Novel REPLI-g SensiPhi DNA Polymerase is used for Multiple Displacement Amplification (MDA). It is a newly developed, high-affinity enzyme that binds cDNA more efficiently, especially when the cDNA concentration is low in the reaction mixture. In addition, in contrast to PCR-based methods, REPLI-g SensiPhi DNA Polymerase has strong proofreading activity that results in 1000-fold fewer errors. It also has strong strand-displacement activity, enabling replication of cDNA through stable hairpin structures that are resistant to Taq-based whole genome or whole transcriptome amplification procedures.
Procedure
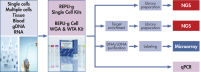
The REPLI-g WTA Single Cell Kit contains reagents for the following sequential reactions (see figure REPLI-g WTA Single Cell Kit procedure):
- Lysis of cells: a single cell sample (containing 1–1000 cells) is lysed efficiently within 5 minutes, with no effect on RNA integrity. The lysed sample is used for WTA of total RNA or, optionally, mRNA-enriched (poly A+) RNA.
- Generation of cDNA: following cell lysis, gDNA is removed prior to the WTA process, since accurate measurement of transcript levels depends on the elimination of false-positive results caused by gDNA contamination. Depending on the primer chosen during the subsequent reverse transcription reaction, all transcripts (if performing total RNA enrichment using random and oligo dT primers) or only poly-adenylated transcripts (if performing poly A+ mRNA enrichment using oligo dT primers) will be amplified. Consequently, the reaction will contain a mixture of random and oligo dT primers, or oligo dT primers only, which reduces rRNA amplification and ensures the 3’ ends of cDNA are reverse transcribed (transcript sizes are approximately 700–1000 bp).
- Ligation: the synthesized cDNA is ligated using a high-efficiency ligation mix. Due to the nature of the subsequent ligation reaction, cDNA fragments are not assembled in the order in which they would have originally existed in the cell. However, this does not affect the detection of nucleic acid sequences, such as splicing regions, in downstream applications like NGS or qPCR.
- Whole transcriptome amplification: The ligated cDNA is amplified utilizing MDA technology, with the novel REPLI-g SensiPhi DNA Polymerase, in an isothermal reaction lasting 2 hours.
Depending on research needs, different protocols are provided for cDNA amplification from single cells or purified RNA:
- The protocol “Amplification of the 3’ Regions of mRNA (Poly A+) from Single Cells” amplifies mRNAs (and other RNAs) with poly A+ tails only and is highly suited for a wide range of applications, including NGS (RNA-Seq), real-time PCR, and microarray analysis.
- The protocol “Amplification of Total RNA from Single Cells” amplifies the complete transcriptome, including RNAs with and without poly A+ tails, lnc RNAs, and linc RNAs. Note that rRNA is also amplified and will be present at a high level following amplification. If working with sequence-specific probes, such as with qPCR or arrays, the amplified rRNA will not affect downstream application results. If using the amplified RNA for RNA-Seq, be aware that more than 90% of reads are derived from rRNA; therefore sufficient reads must be obtained when performing whole-transcriptome sequencing.
- The protocol “Amplification of Purified RNA” is optimized for whole transcriptome amplification from total or enriched RNA templates and is highly suited for a wide range of applications, including NGS, real-time PCR, and microarray analysis.
Applications
The REPLI-g WTA Single Cell Kit allows uniform amplification of all transcripts from very small samples, accurately representing the transcription pattern of a single cell with very limited, or no, amplification bias. Depending on the protocol, amplified cDNA is highly suited for use in next-generation sequencing (RNA-Seq), gene expression arrays, or for quantitative PCR analysis.
The REPLI-g WTA Single Cell Kit can be used for whole transcriptome amplification for analysis of:
- mRNA with poly A+ tails
- Total RNA
- All regions of RNA transcripts
- 3’ ends of mRNAs
- Lnc and linc RNAs
It is not suitable for use with small nucleic acids (for example):
- tRNAs and miRNAs
Supporting data and figures
High number of mappable reads from just 3 cells.